Article Search
Volume 27.1
January–April 2024
Full table of contents
ISSN: 1094-8074, web version;
1935-3952, print version
Recent Research Articles
See all articles in 27.1 January-April 2024
See all articles in 26.3 September-December 2023
See all articles in 26.2 May-August 2023
See all articles in 26.1 January-April 2023
 Matthijs Freudenthal. Departamento de Estratigrafía y Paleontología, Universidad de Granada, Avda. Fuentenueva s/n, E-18071 Granada, Spain; Naturalis Biodiversity Center, P.O. Box 9517, NL-2300 RA Leiden, The Netherlands. mfreuden@ugr.es
Matthijs Freudenthal. Departamento de Estratigrafía y Paleontología, Universidad de Granada, Avda. Fuentenueva s/n, E-18071 Granada, Spain; Naturalis Biodiversity Center, P.O. Box 9517, NL-2300 RA Leiden, The Netherlands. mfreuden@ugr.es
Matthijs Freudenthal is a former curator of vertebrate paleontology of the National Museum of Natural History, Naturalis, Leiden, the Netherlands. He is actually working at the University of Granada, Spain. He is one of the pioneers of fossil micromammal research in Europe. His most recent research covers a variety of subjects like taxonomy of Gliridae, correspondance analysis in vertebrate paleontology, the insular faunas from Gargano (Italy) and body mass estimates in fossil rodents.

 Elvira Martín-Suárez. Departamento de Estratigrafía y Paleontología, Universidad de Granada, Avda. Fuentenueva s/n, E-18071 Granada, Spain. elvirams@ugr.es
Elvira Martín-Suárez. Departamento de Estratigrafía y Paleontología, Universidad de Granada, Avda. Fuentenueva s/n, E-18071 Granada, Spain. elvirams@ugr.es
Elvira Martín Suárez is professor of vertebrate paleontology at the University of Granada, Spain. After completing her study in Biology, she specialized in the field of fossil vertebrates at the University of Lyon (France) and the National Museum of Natural History, Leiden, the Netherlands. Her main scientific interest are the fossil rodents from the Mio-Pliocene transition.
TABLE 1. Species of Plesiosminthus described from the European Oligocene and Miocene.
| Species | Type-locality |
| P. schaubi Viret, 1926 (type-species) | Coderet, France |
| P. myarion Schaub, 1930 | Chavroches, France |
| P. promyarion Schaub, 1930 | Puy-de-Montdoury, F rance |
| P. bavaricus Freudenberg, 1941 | Gaimersheim, Germany |
| P. winistoerferi Engesser, 1987 | Brochene Fluh 53, Switzerland |
| P. conjunctus Ziegler, 1994 | Herrlingen 8, Germany |
| P. moralesi Álvarez Sierra, Daams and Lacomba Andueza, 1996 | Sayatón 1, Spain |
| P. admyarion Comte, 2000 | Thézels, France |
| P. meridionalis Comte, 2000 | Venelles inf., France |
| P. margaritae n. sp. (this paper) | Mirambueno 1, Spain |
| P. moniqueae n. sp. (this paper) | Pech Desse, France |
| Plesiosminthus n. sp., Ziegler, 1994 | Herrlingen 8, Germany |
| Plesiosminthus sp., Engesser and Mödden, 1987 | Mümliswil-Hardberg, Switzerland |
| Plesiosminthus sp., Comte, 2000 | Venelles inf., France |
TABLE 2. Locality codes, species contents and most important reference of the localities studied. Zones are European mammal zones (Biochrom’97, 1997).
| Code | Locality | Zone | Species | Reference |
| BF53 | Brochene Fluh 53 | MP30 | P. winistoerferi | Engesser (1987) |
| CANAL | Canales | MP28 | Plesiosminthus sp. | Alvarez Sierra et al. (1999) |
| CHAVR | Chavroches | MN2 | P. myarion | Hugueney and Vianey-Liaud (1980) |
| CLUZEL | Cluzel | MN2 | P. myarion | Hugueney and Vianey-Liaud (1980) |
| COD3 | Coderet 3 | MP30 | P. schaubi | Hugueney (1969) |
| DIEU | Dieupentale | MP30 | P. schaubi | Baudelot and Olivier (1978) |
| FORN11 | Fornant 11 | MN1 | P. myarion | Engesser (1987) |
| GAIM | Gaimersheim | MP28 | P. bavaricus | Kristkoiz (1992) |
| HERR8 | Herrlingen 8 | MP28 | P. conjunctus | Ziegler (1994) |
| LAUT2 | Lautern 2 | MN1 | P. myarion | Ziegler and Werner (1994) |
| MIR1 | Mirambueno 1 | MP27 | P. margaritae | this paper |
| MIR2A | Mirambueno 2A | MP27 | P. aff. conjunctus | this paper |
| OBL | Oberleichtersbach | MP30 | P. winistoerferi | Engesser and Storch (2008) |
| PARR | Parrales | MP29 | P. schaubi | Alvarez Sierra et al. (1999) |
| PDES | Pech Desse | MP28 | P. promyarion | Hugueney and Vianey-Liaud (1980) |
| PDES | Pech Desse | MP28 | P. moniqueae | this paper |
| Pech-du-Fraysse | MP28 | P. promyarion | Hugueney and Vianey-Liaud (1980) | |
| RDUBEY | Ruisseau du Bey | MP28 | P. promyarion | Engesser (1987) |
| SAULC | Saulcet | MN1 | P. myarion | Hugueney and Vianey-Liaud (1980) |
| SAY1 | Sayatón 1 | MP29 | P. moralesi | Alvarez et al. (1996) |
| THZ | Thézels | MP30 | P. admyarion | Comte (2000) |
| VENINF | Venelles inf. | MP30 | P. meridionalis | Comte (2000) |
| VIV1 | Vivel del Río 1 | MP28 | P. cf. margaritae | this paper |
TABLE 3. Frequencies of character states of the protolophule of M2, number of observations, and morphology values. Ant. = anterior, trans = transverse, ant.plus = anterior plus an incomplete posterior connection. * protolophule posterior, not double.
| Sample | ant. or trans | ant. plus | double | n | MV |
| CHAVR | 0 | 3 | 97 | 33 | 0.985 |
| FORN11 | |||||
| VENINF | 4 | 0 | 96* | 25 | 0.960 |
| THZ | 95 | 0 | 5 | 97 | 0.050 |
| COD3 | 97 | 0 | 3 | 63 | 0.032 |
| HERR8 | 17 | 8 | 75 | 12 | 0.790 |
| GAIM | 39 | 52 | 9 | 23 | 0.350 |
| PDES | 74 | 13 | 13 | 16 | 0.195 |
| RDUBEY | |||||
| VIV1 | 57 | 4 | 39 | 69 | 0.410 |
| MIR2A | 19 | 7 | 74 | 27 | 0.775 |
| MIR1 | 28 | 37 | 35 | 43 | 0.535 |
TABLE 4. P-values of t-test comparing the samples from THZ and COD3; df = degrees of freedom.
| Molar | Comte (2000) |
this paper | df |
| m1 | 3.18 | 3.092 | 205 |
| m2 | 2.02 | -2.162 | 180 |
| m3 | 3.00 | 2.828 | 127 |
| M1 | 2.39 | -2.483 | 188 |
| M2 | 2.06 | -2.178 | 172 |
| M3 | 3.00 | 2.832 | 98 |
TABLE 5. Frequencies of character states of the protoconid hind arm of m2, number of observations, and morphology values. Data from Kristkoiz (1992, figure 84) and this paper.
| Sample | complete | incomplete | absent | n | MV |
| CHAVR | 12 | 50 | 38 | 50 | 0.630 |
| FORN11 | 26 | 52 | 22 | 27 | 0.480 |
| VENINF | 0 | 0 | 100 | 26 | 1.000 |
| COD3 | 0 | 0 | 100 | 59 | 1.000 |
| THZ | 9 | 9 | 82 | 127 | 0.865 |
| HERR8 | 82 | 18 | 0 | 17 | 0.090 |
| 36 | 53 | 11 | 28 | 0.375 | |
| GAIM | 50 | 41 | 9 | 31 | 0.295 |
| PDES | 32 | 45 | 23 | 53 | 0.455 |
| RDUBEY | 31 | 69 | 0 | 13 | 0.345 |
| VIV1 | 31 | 16 | 53 | 52 | 0.610 |
| MIR2A | 83 | 0 | 17 | 16 | 0.170 |
| MIR1 | 60 | 26 | 14 | 41 | 0.270 |
TABLE 6. Frequencies of character states of the protoconid hind arm of m3, number of observations, and morphology values.
| Sample | complete | incomplete | absent | n | MV |
| CHAVR | 5.4 | 0.0 | 94.6 | 37 | 0.946 |
| VENINF | 0.0 | 0.0 | 100.0 | 8 | 1.000 |
| COD3 | 0.0 | 0.0 | 100.0 | 32 | 1.000 |
| THZ | 0.0 | 0.0 | 100.0 | 97 | 1.000 |
| HERR8 | 0.0 | 0.0 | 100.0 | 14 | 1.000 |
| 8.0 | 32.0 | 60.0 | 25 | 0.760 | |
| GAIM | 14.9 | 27.3 | 63.6 | 11 | 0.730 |
| PDES | 25.0 | 43.8 | 31.2 | 16 | 0.531 |
| RDUBEY | 0.0 | 0.0 | 100.0 | 6 | 1.000 |
| VIV1 | 5.1 | 0.0 | 94.9 | 39 | 0.949 |
| MIR2A | 20.0 | 15.0 | 65.0 | 20 | 0.725 |
| MIR1 | 23.2 | 11.6 | 65.1 | 43 | 0.710 |
TABLE 7. Number of specimens for samples in Figure 13.
| Sample | m1 | m2 | m3 | M1 | M2 | M3 |
| BF53 | 4 | 4 | 5 | 6 | 5 | 5 |
| CHAVR | 36 | 48 | 39 | 49 | 33 | 15 |
| COD3 | 65 | 59 | 32 | 71 | 63 | 13 |
| GAIM | 34 | 32 | 11 | 28 | 23 | 3 |
| HERR8 | 8 | 15 | 14 | 18 | 12 | 11 |
| MIR1 | 17 | 41 | 45 | 49 | 43 | 37 |
| PDES | 57 | 54 | 16 | 35 | 16 | 3 |
| SAY1 | 7 | 6 | 1 | 3 | 3 | 1 |
| THZ | 142 | 123 | 97 | 119 | 111 | 87 |
| VENINF | 19 | 25 | 8 | 27 | 26 | 1 |
FIGURE 1. Terminology of molars. Figures represent left-hand molars.
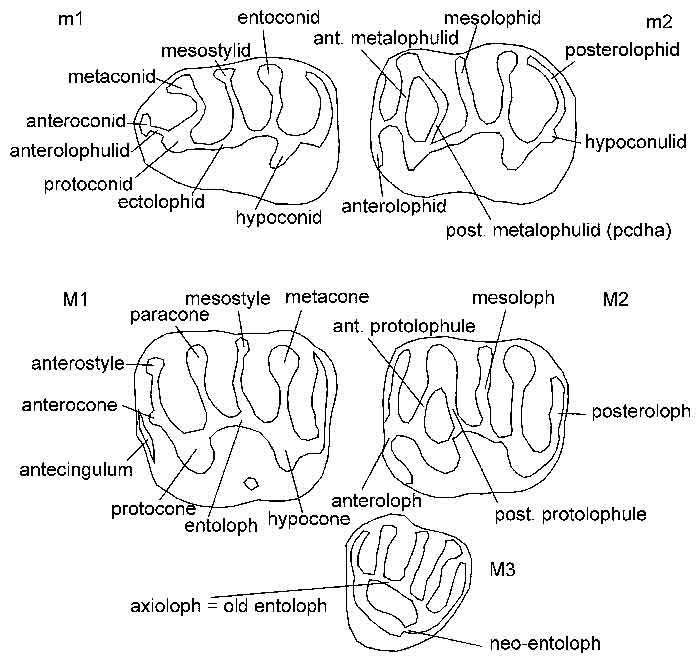
FIGURE 2. Distribution of morphology values of protoconid hind arm in m2 (filled circles) and protolophule in M2 (open circles). Enclosing circles and connecting lines indicate possible relationships.
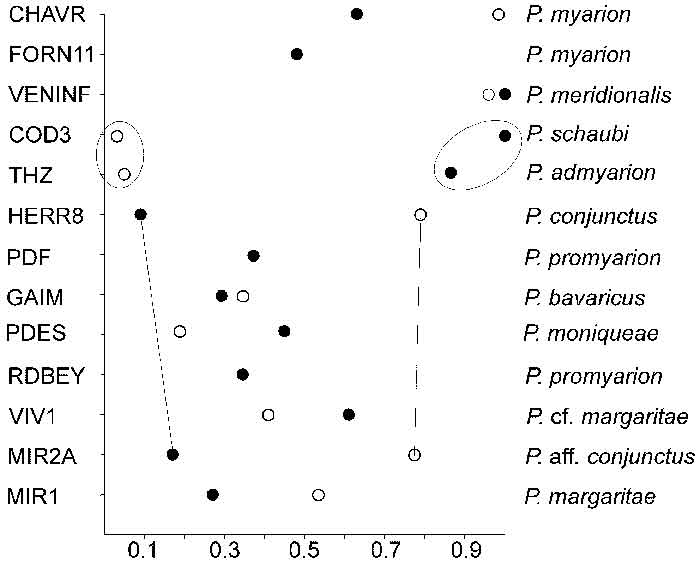
FIGURE 3. Plesiosminthus margaritae n. sp. from Mirambueno 1 (occlusal views). 1, m1 sin., RGM 558099; 2, m2 sin., MIR1 168; 3, m3 sin., RGM 558149; 4, M1 sin., RGM 558200; 5, M2 sin., MIR1 189; 6, M3 sin., MIR1 196; 7, m1 dext., RGM 558105; 8, m2 dext., RGM 558138; 9, m3 dext., MIR1 178; 10, M1 dext., MIR1 187; 11, M2 dext., MIR1 193 (Holotype); and 12, M3 dext., MIR1 197.
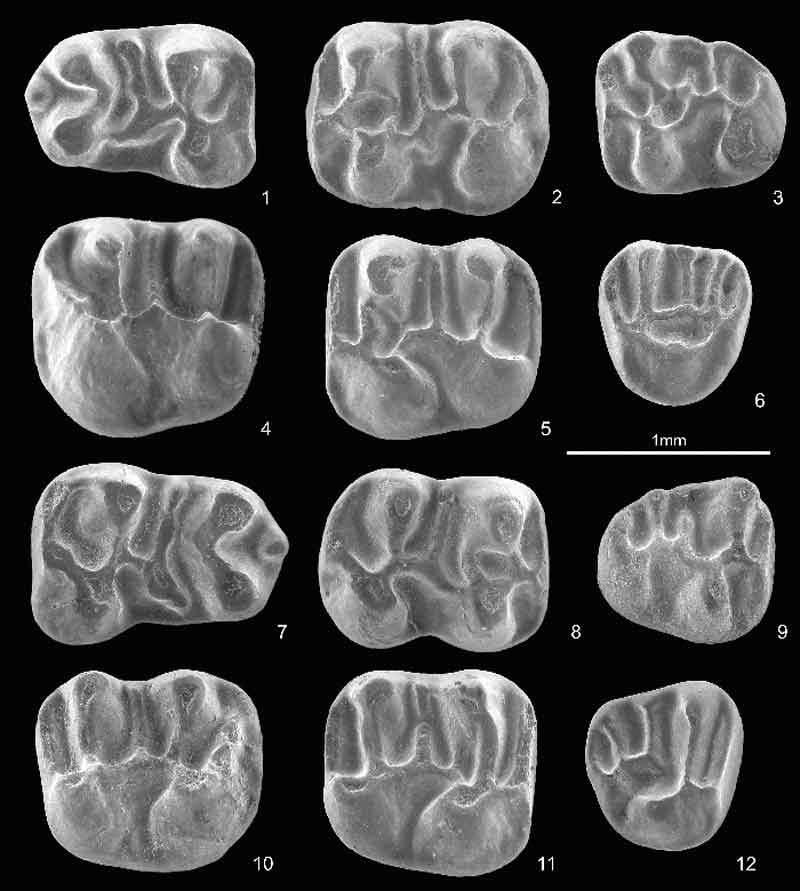
FIGURE 4. Scatter diagrams of molar length (L) and width (W) of Plesiosminthus margaritae n. sp. from Mirambueno 1 (in mm). 1, m1; 2, m2; 3, m3; 4, M1; 5, M2; and 6, M3.
FIGURE 5. Scatter diagrams of molar length (L) and width (W) of Plesiosminthus cf. margaritae n. sp. from Vivel del Río 1 (in mm). 1, m1; 2, m2; 3, m3; 4, M1; 5, M2; and 6, M3.
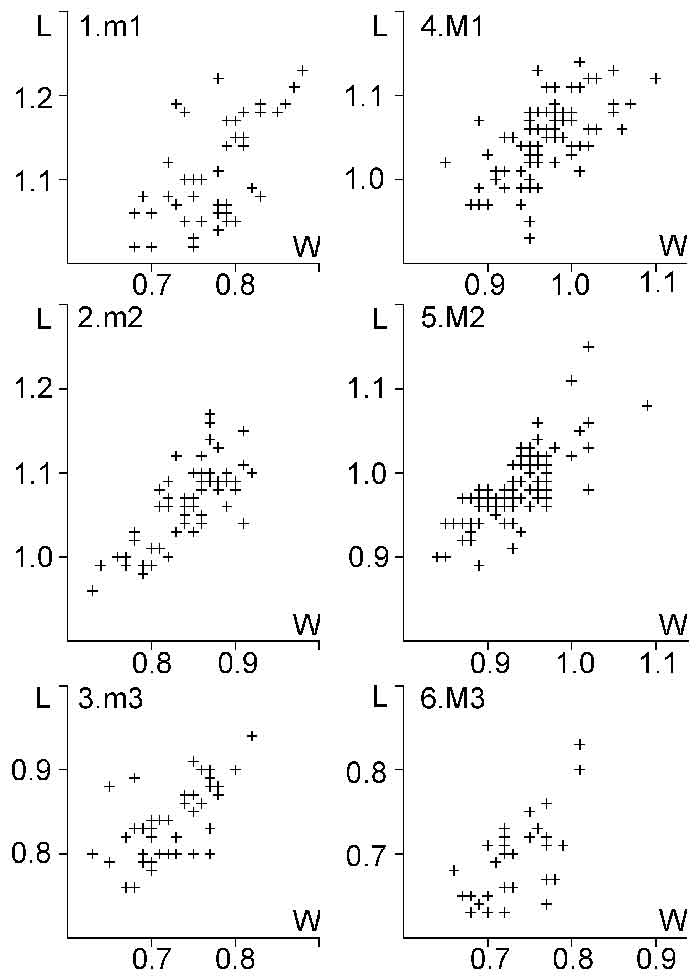
FIGURE 6. Scatter diagrams of molar length (L) and width (W) of Plesiosminthus aff. conjunctus (cross) and Plesiosminthus sp. (circle) from Mirambueno 2A, compared with the distribution area (rectangle) of Plesiosminthus conjunctus molars from HERR8 (in mm). 1, m1; 2, m2; 3, m3; 4, M1; 5, M2; and 6, M3.
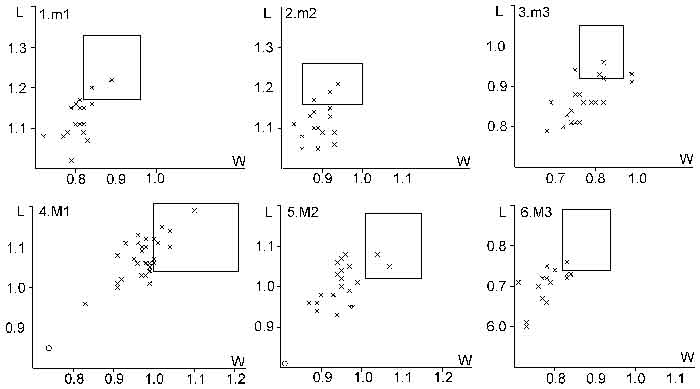
FIGURE 7. m1 length (1) and width (2) of Plesiosminthus species. The bars represent the minimum and maximum in mm, a tick marks the mean, and the number of specimens is given to the right.
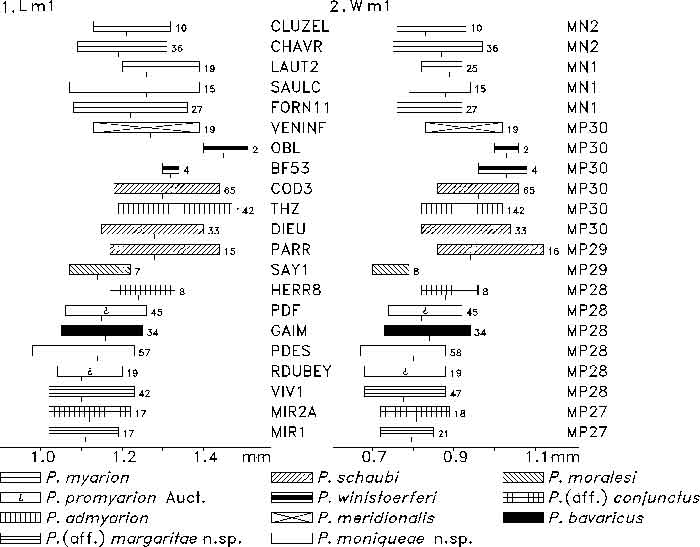
FIGURE 8. m2 length (1) and width (2) of Plesiosminthus species. The bars represent the minimum and maximum in mm, a tick marks the mean, and the number of specimens is given to the right.
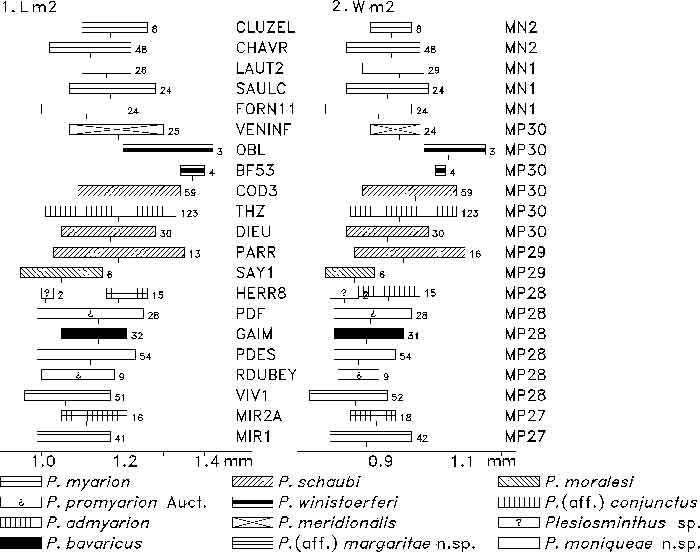
FIGURE 9. m3 length (1) and width (2) of Plesiosminthus species. The bars represent the minimum and maximum in mm, a tick marks the mean, and the number of specimens is given to the right.
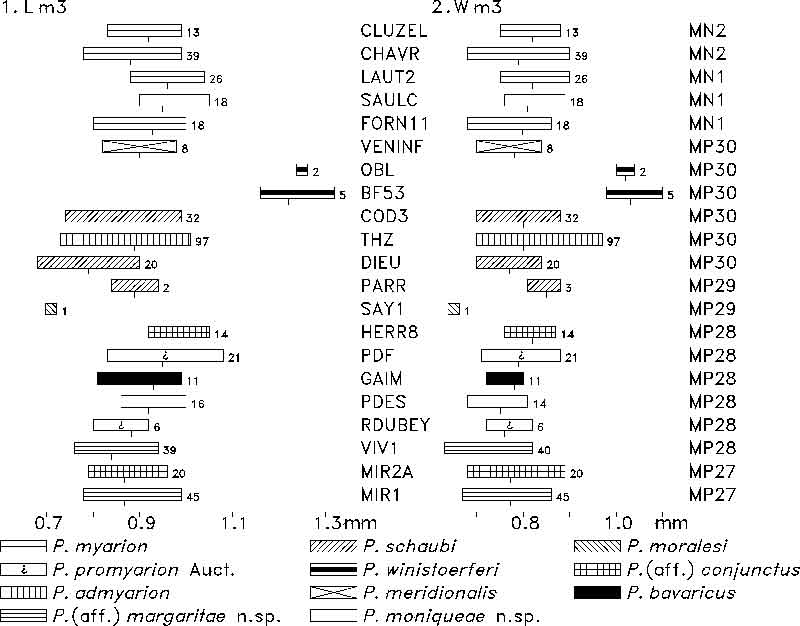
FIGURE 10. M1 length (1) and width (2) of Plesiosminthus species. The bars represent the minimum and maximum in mm, a tick marks the mean, and the number of specimens is given to the right.
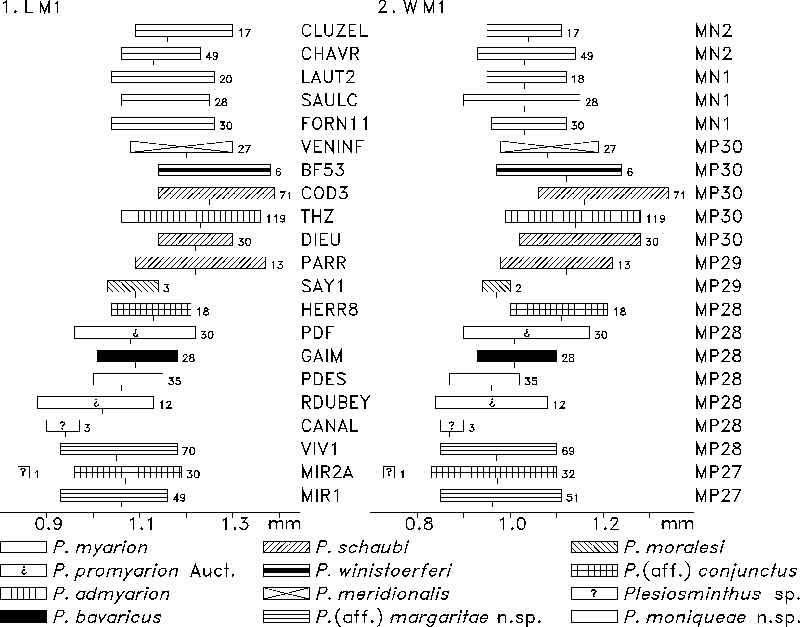
FIGURE 11. M2 length (1) and width (2) of Plesiosminthus species. The bars represent the minimum and maximum in mm, a tick marks the mean, and the number of specimens is given to the right.
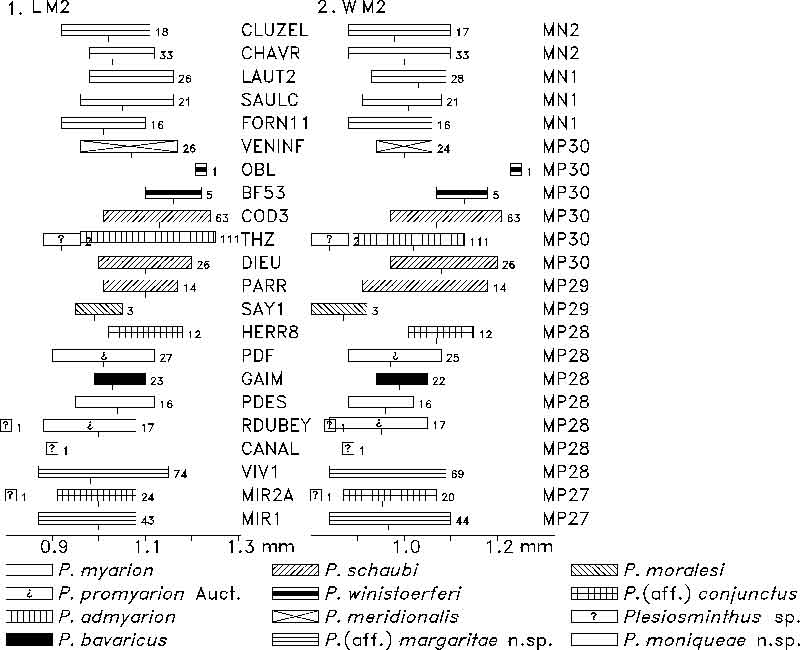
FIGURE 12. M3 length (1) and width (2) of Plesiosminthus species. The bars represent the minimum and maximum in mm, a tick marks the mean, and the number of specimens is given to the right.
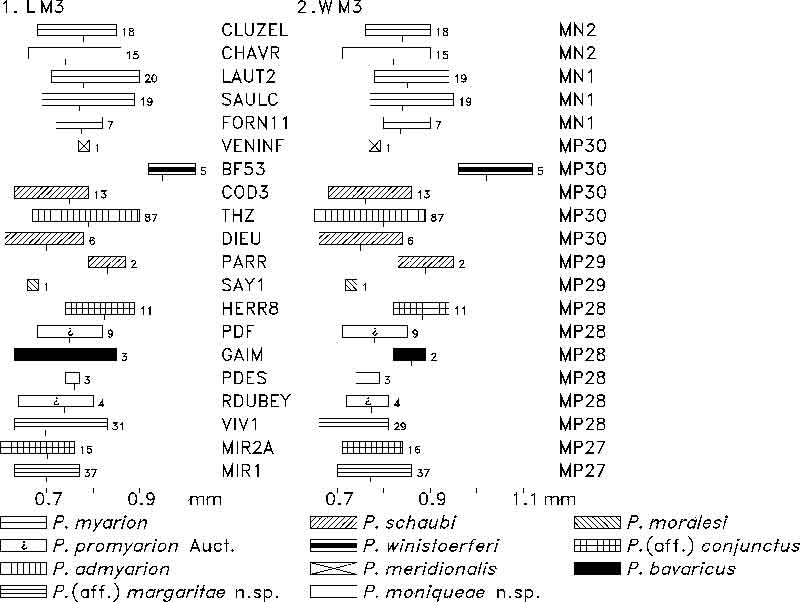
FIGURE 13. Relation of mean length values (in mm) of m1, m2, m3, M1, M2, M3 of Plesiosminthus species. Number of specimens per sample in Table 7.
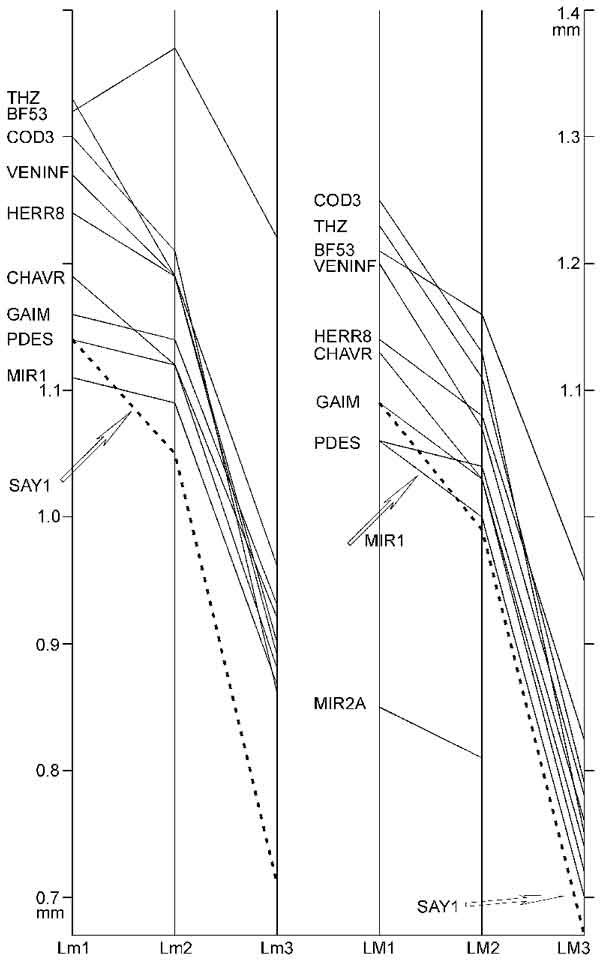
Measurements of Plesiosminthus samples. V' = coefficient of variation as defined by Freudenthal and Cuenca Bescos (1984): 100R/M, where R is the range, and M is the median of the distribution. σ = standard deviation.
| Length | Width | ||||||||||||
| m1 | N | Min. | Mean | Max. | V' | σ | N | Min. | Mean | Max. | V' | σ | |
| CLUZEL | 10 | 1.13 | 1.210 | 1.32 | 15.5 | 0.055 | 10 | 0.76 | 0.830 | 0.93 | 20.1 | 0.046 | |
| CHAVR | 36 | 1.09 | 1.190 | 1.31 | 18.3 | 0.013 | 36 | 0.75 | 0.870 | 0.97 | 25.6 | 0.049 | |
| LAUT2 | 19 | 1.20 | 1.260 | 1.39 | 14.7 | 0.053 | 25 | 0.82 | 0.890 | 0.92 | 11.5 | 0.042 | |
| SAULC | 15 | 1.07 | 1.260 | 1.39 | 26.0 | 0.059 | 15 | 0.79 | 0.880 | 0.94 | 17.3 | 0.044 | |
| FORN11 | 27 | 1.08 | 1.222 | 1.36 | 23.0 | 0.063 | 27 | 0.76 | 0.856 | 0.92 | 19.0 | 0.042 | |
| VENINF | 19 | 1.13 | 1.270 | 1.39 | 20.6 | 0.064 | 19 | 0.83 | 0.910 | 1.02 | 20.5 | 0.058 | |
| BF53 | 4 | 1.30 | 1.320 | 1.34 | 3.0 | 0.016 | 4 | 0.96 | 1.035 | 1.08 | 11.8 | 0.054 | |
| THZ | 142 | 1.19 | 1.330 | 1.47 | 21.1 | 0.062 | 142 | 0.82 | 0.920 | 1.02 | 21.7 | 0.045 | |
| COD3 | 65 | 1.18 | 1.300 | 1.44 | 19.8 | 0.066 | 65 | 0.86 | 0.960 | 1.06 | 20.8 | 0.046 | |
| DIEU | 33 | 1.15 | 1.28 | 1.40 | 19.6 | 33 | 0.82 | 0.93 | 1.04 | 23.7 | |||
| PARR | 15 | 1.17 | 1.280 | 1.44 | 20.7 | 16 | 0.86 | 0.940 | 1.12 | 26.3 | |||
| SAY1 | 7 | 1.07 | 1.140 | 1.22 | 13.1 | 8 | 0.70 | 0.750 | 0.79 | 12.1 | |||
| HERR8 | 8 | 1.17 | 1.240 | 1.33 | 12.8 | 8 | 0.82 | 0.879 | 0.96 | 15.7 | |||
| 45 | 1.06 | 1.150 | 1.26 | 17.2 | 0.049 | 45 | 0.74 | 0.820 | 0.92 | 21.7 | 0.043 | ||
| PDES | 57 | 0.98 | 1.140 | 1.23 | 22.6 | 0.049 | 58 | 0.67 | 0.800 | 0.88 | 27.1 | 0.039 | |
| GAIM | 34 | 1.05 | 1.160 | 1.25 | 17.4 | 0.051 | 34 | 0.73 | 0.840 | 0.94 | 25.1 | 0.051 | |
| RDUBEY | 19 | 1.04 | 1.109 | 1.20 | 14.3 | 0.051 | 19 | 0.68 | 0.792 | 0.88 | 25.6 | 0.057 | |
| VIV1 | 42 | 1.02 | 1.109 | 1.23 | 18.7 | 0.062 | 47 | 0.68 | 0.776 | 0.88 | 25.6 | 0.050 | |
| MIR2A | 17 | 1.02 | 1.125 | 1.22 | 17.9 | 0.051 | 18 | 0.72 | 0.808 | 0.89 | 21.1 | 0.035 | |
| MIR1 | 17 | 1.02 | 1.119 | 1.19 | 15.4 | 0.056 | 21 | 0.72 | 0.796 | 0.85 | 16.6 | 0.039 | |
|
|
|||||||||||||
| m2 | N | Min. | Mean | Max. | V' | σ | N | Min. | Mean | Max. | V' | σ | |
| CLUZEL | 8 | 1.10 | 1.170 | 1.26 | 13.6 | 0.058 | 8 | 0.88 | 0.930 | 0.98 | 10.8 | 0.033 | |
| CHAVR | 48 | 1.02 | 1.120 | 1.22 | 17.9 | 0.050 | 48 | 0.82 | 0.930 | 1.00 | 19.8 | 0.041 | |
| LAUT2 | 28 | 1.10 | 1.160 | 1.22 | 10.3 | 0.034 | 29 | 0.86 | 0.950 | 1.01 | 16.0 | 0.048 | |
| SAULC | 24 | 1.07 | 1.170 | 1.28 | 17.9 | 0.057 | 24 | 0.82 | 0.920 | 1.02 | 21.7 | 0.050 | |
| FORN11 | 24 | 1.00 | 1.116 | 1.20 | 18.2 | 0.052 | 24 | 0.77 | 0.898 | 0.98 | 24.0 | 0.054 | |
| VENINF | 25 | 1.07 | 1.190 | 1.30 | 19.4 | 0.049 | 24 | 0.88 | 0.950 | 1.00 | 12.8 | 0.041 | |
| BF53 | 4 | 1.34 | 1.373 | 1.40 | 4.4 | 0.032 | 4 | 1.04 | 1.048 | 1.06 | 1.9 | 0.010 | |
| THZ | 123 | 1.01 | 1.190 | 1.33 | 27.4 | 0.065 | 123 | 0.83 | 0.950 | 1.09 | 27.1 | 0.051 | |
| COD3 | 59 | 1.09 | 1.210 | 1.34 | 20.6 | 0.055 | 59 | 0.86 | 0.990 | 1.09 | 23.6 | 0.049 | |
| DIEU | 30 | 1.05 | 1.17 | 1.28 | 19.7 | 30 | 0.82 | 0.92 | 1.02 | 21.7 | |||
| PARR | 13 | 1.03 | 1.190 | 1.35 | 26.9 | 16 | 0.84 | 0.960 | 1.11 | 27.7 | |||
| SAY1 | 6 | 0.95 | 1.050 | 1.15 | 19.0 | 6 | 0.77 | 0.840 | 0.89 | 14.5 | |||
| HERR8 | 15 | 1.16 | 1.197 | 1.26 | 8.3 | 15 | 0.85 | 0.923 | 1.00 | 16.2 | |||
| 28 | 0.99 | 1.140 | 1.25 | 23.2 | 0.067 | 28 | 0.79 | 0.880 | 0.98 | 21.5 | 0.047 | ||
| PDES | 54 | 0.99 | 1.120 | 1.23 | 21.6 | 0.048 | 54 | 0.79 | 0.850 | 0.94 | 17.3 | 0.038 | |
| GAIM | 32 | 1.05 | 1.140 | 1.21 | 14.2 | 0.041 | 31 | 0.79 | 0.870 | 0.96 | 19.4 | 0.039 | |
| RDUBEY | 9 | 1.00 | 1.106 | 1.18 | 16.5 | 0.066 | 9 | 0.80 | 0.841 | 0.90 | 11.8 | 0.044 | |
| VIV1 | 51 | 0.96 | 1.063 | 1.17 | 19.7 | 0.050 | 52 | 0.73 | 0.842 | 0.92 | 23.0 | 0.046 | |
| MIR2A | 16 | 1.05 | 1.116 | 1.21 | 14.2 | 0.048 | 18 | 0.83 | 0.893 | 0.94 | 12.4 | 0.032 | |
| MIR1 | 41 | 0.99 | 1.093 | 1.17 | 16.7 | 0.041 | 42 | 0.78 | 0.869 | 0.98 | 22.7 | 0.041 | |
|
|
|||||||||||||
| m3 | N | Min. | Mean | Max. | V' | σ | N | Min. | Mean | Max. | V' | σ | |
| CLUZEL | 13 | 0.83 | 0.920 | 0.99 | 17.6 | 0.044 | 13 | 0.75 | 0.820 | 0.88 | 16.0 | 0.043 | |
| CHAVR | 39 | 0.78 | 0.880 | 0.99 | 23.7 | 0.056 | 39 | 0.68 | 0.790 | 0.90 | 27.8 | 0.047 | |
| LAUT2 | 26 | 0.88 | 0.960 | 1.04 | 16.7 | 0.051 | 26 | 0.75 | 0.820 | 0.90 | 18.2 | 0.044 | |
| SAULC | 18 | 0.90 | 0.950 | 1.05 | 15.4 | 0.036 | 18 | 0.76 | 0.810 | 0.89 | 15.8 | 0.040 | |
| FORN11 | 18 | 0.80 | 0.928 | 1.00 | 22.2 | 0.042 | 18 | 0.68 | 0.799 | 0.86 | 23.4 | 0.043 | |
| VENINF | 8 | 0.82 | 0.900 | 0.98 | 17.8 | 0.052 | 8 | 0.70 | 0.780 | 0.84 | 18.2 | 0.046 | |
| BF53 | 5 | 1.16 | 1.220 | 1.32 | 12.9 | 0.062 | 5 | 0.98 | 1.032 | 1.10 | 11.5 | 0.063 | |
| THZ | 97 | 0.73 | 0.890 | 1.01 | 32.2 | 0.049 | 97 | 0.70 | 0.800 | 0.97 | 32.3 | 0.052 | |
| COD3 | 32 | 0.74 | 0.860 | 0.99 | 28.9 | 0.053 | 32 | 0.70 | 0.800 | 0.88 | 22.8 | 0.045 | |
| DIEU | 20 | 0.68 | 0.79 | 0.90 | 27.8 | 20 | 0.70 | 0.77 | 0.84 | 18.2 | |||
| PARR | 2 | 0.84 | 0.890 | 0.94 | 11.2 | 3 | 0.81 | 0.850 | 0.88 | 8.3 | |||
| SAY1 | 1 | 0.710 | 1 | 0.650 | |||||||||
| HERR8 | 14 | 0.92 | 0.961 | 1.05 | 13.2 | 14 | 0.76 | 0.819 | 0.87 | 13.5 | |||
| 21 | 0.83 | 0.950 | 1.08 | 26.2 | 0.057 | 21 | 0.71 | 0.790 | 0.88 | 21.4 | 0.046 | ||
| PDES | 16 | 0.86 | 0.920 | 1.00 | 15.1 | 0.041 | 14 | 0.68 | 0.750 | 0.81 | 17.4 | 0.037 | |
| GAIM | 11 | 0.81 | 0.930 | 0.99 | 20.0 | 0.050 | 11 | 0.72 | 0.780 | 0.80 | 10.5 | 0.024 | |
| RDUBEY | 6 | 0.80 | 0.883 | 0.92 | 14.0 | 0.043 | 6 | 0.72 | 0.760 | 0.82 | 13.0 | 0.044 | |
| VIV1 | 39 | 0.76 | 0.839 | 0.94 | 21.2 | 0.045 | 40 | 0.63 | 0.724 | 0.82 | 26.2 | 0.044 | |
| MIR2A | 20 | 0.79 | 0.867 | 0.96 | 19.4 | 0.051 | 20 | 0.68 | 0.773 | 0.89 | 26.8 | 0.056 | |
| MIR1 | 45 | 0.78 | 0.867 | 0.99 | 23.7 | 0.046 | 45 | 0.67 | 0.773 | 0.86 | 24.8 | 0.045 | |
|
|
|||||||||||||
| P4 | N | Min. | Mean | Max. | V' | σ | N | Min. | Mean | Max. | V' | σ | |
| CLUZEL | 7 | 0.47 | 0.550 | 0.60 | 24.3 | 0.042 | 7 | 0.49 | 0.560 | 0.65 | 28.1 | 0.038 | |
| CHAVR | 18 | 0.36 | 0.470 | 0.60 | 50.0 | 0.051 | 18 | 0.43 | 0.530 | 0.65 | 40.7 | 0.049 | |
| SAULC | 21 | 0.40 | 0.480 | 0.58 | 36.7 | 0.053 | 21 | 0.42 | 0.520 | 0.64 | 41.5 | 0.054 | |
| THZ | 34 | 0.47 | 0.610 | 0.71 | 40.7 | 0.043 | 34 | 0.57 | 0.650 | 0.73 | 24.6 | 0.035 | |
| COD3 | 29 | 0.48 | 0.590 | 0.66 | 31.6 | 0.040 | 29 | 0.51 | 0.620 | 0.73 | 35.5 | 0.049 | |
| GAIM | 1 | 0.500 | 1 | 0.600 | |||||||||
| VIV1 | 7 | 0.49 | 0.536 | 0.59 | 18.5 | 0.036 | 7 | 0.52 | 0.559 | 0.60 | 14.3 | 0.027 | |
| MIR2A | 2 | 0.56 | 0.565 | 0.57 | 1.8 | 2 | 0.47 | 0.505 | 0.54 | 13.9 | |||
| MIR1 | 1 | 0.540 | 1 | 0.590 | |||||||||
|
|
|||||||||||||
| M1 | N | Min. | Mean | Max. | V' | σ | N | Min. | Mean | Max. | V' | σ | |
| CLUZEL | 17 | 1.09 | 1.160 | 1.30 | 17.6 | 0.053 | 17 | 0.95 | 1.040 | 1.11 | 15.5 | 0.043 | |
| CHAVR | 49 | 1.06 | 1.130 | 1.23 | 14.8 | 0.044 | 49 | 0.93 | 1.030 | 1.14 | 20.3 | 0.056 | |
| LAUT2 | 20 | 1.04 | 1.150 | 1.26 | 19.1 | 0.054 | 18 | 0.95 | 1.030 | 1.12 | 16.4 | 0.049 | |
| SAULC | 28 | 1.06 | 1.150 | 1.25 | 16.5 | 0.047 | 28 | 0.90 | 1.030 | 1.15 | 24.4 | 0.055 | |
| FORN11 | 30 | 1.04 | 1.150 | 1.26 | 19.1 | 0.050 | 30 | 0.96 | 1.036 | 1.12 | 15.4 | 0.053 | |
| VENINF | 27 | 1.08 | 1.200 | 1.30 | 18.5 | 0.049 | 27 | 0.98 | 1.080 | 1.19 | 19.4 | 0.049 | |
| BF53 | 6 | 1.14 | 1.217 | 1.38 | 19.0 | 0.089 | 6 | 0.97 | 1.122 | 1.24 | 24.4 | 0.088 | |
| THZ | 119 | 1.06 | 1.230 | 1.36 | 24.8 | 0.058 | 119 | 0.99 | 1.140 | 1.28 | 25.6 | 0.059 | |
| COD3 | 71 | 1.14 | 1.250 | 1.39 | 19.8 | 0.051 | 71 | 1.06 | 1.160 | 1.34 | 23.3 | 0.052 | |
| DIEU | 30 | 1.14 | 1.22 | 1.30 | 13.1 | 30 | 1.02 | 1.15 | 1.28 | 22.6 | |||
| PARR | 13 | 1.09 | 1.220 | 1.37 | 22.8 | 13 | 0.98 | 1.120 | 1.22 | 21.8 | |||
| SAY1 | 3 | 1.03 | 1.090 | 1.14 | 10.1 | 2 | 0.94 | 0.970 | 1.00 | 6.2 | |||
| HERR8 | 18 | 1.04 | 1.139 | 1.21 | 15.1 | 18 | 1.00 | 1.111 | 1.21 | 19.0 | |||
| 30 | 0.96 | 1.080 | 1.22 | 23.9 | 0.067 | 30 | 0.90 | 1.010 | 1.17 | 26.1 | 0.065 | ||
| PDES | 35 | 1.00 | 1.060 | 1.15 | 14.0 | 0.035 | 35 | 0.87 | 0.960 | 1.02 | 15.9 | 0.035 | |
| GAIM | 28 | 1.01 | 1.090 | 1.18 | 15.5 | 0.044 | 28 | 0.93 | 1.010 | 1.10 | 16.7 | 0.045 | |
| RDUBEY | 12 | 0.88 | 1.023 | 1.13 | 24.9 | 0.088 | 12 | 0.84 | 0.948 | 1.08 | 25.0 | 0.080 | |
| VIV1 | 70 | 0.93 | 1.050 | 1.18 | 23.7 | 0.049 | 69 | 0.85 | 0.970 | 1.10 | 25.6 | 0.048 | |
| MIR2A | 30 | 0.96 | 1.075 | 1.19 | 21.4 | 0.050 | 32 | 0.83 | 0.973 | 1.10 | 28.0 | 0.049 | |
| MIR1 | 49 | 0.93 | 1.062 | 1.16 | 22.0 | 0.048 | 51 | 0.85 | 0.962 | 1.11 | 26.5 | 0.051 | |
|
|
|||||||||||||
| M2 | N | Min. | Mean | Max. | V' | σ | N | Min. | Mean | Max. | V' | σ | |
| CLUZEL | 18 | 0.92 | 1.020 | 1.11 | 18.7 | 0.052 | 17 | 0.88 | 0.980 | 1.10 | 22.2 | 0.054 | |
| CHAVR | 33 | 0.98 | 1.030 | 1.12 | 13.3 | 0.057 | 33 | 0.88 | 1.000 | 1.10 | 22.2 | 0.057 | |
| LAUT2 | 26 | 0.98 | 1.080 | 1.16 | 16.8 | 0.047 | 28 | 0.93 | 1.030 | 1.09 | 15.8 | 0.049 | |
| SAULC | 21 | 0.96 | 1.050 | 1.16 | 18.9 | 0.051 | 21 | 0.91 | 1.010 | 1.08 | 17.1 | 0.048 | |
| FORN11 | 16 | 0.92 | 1.016 | 1.10 | 17.8 | 0.055 | 16 | 0.88 | 0.959 | 1.06 | 18.6 | 0.058 | |
| VENINF | 26 | 0.96 | 1.070 | 1.17 | 19.7 | 0.057 | 24 | 0.94 | 1.000 | 1.06 | 12.0 | 0.036 | |
| BF53 | 5 | 1.10 | 1.164 | 1.22 | 10.3 | 0.048 | 5 | 1.07 | 1.134 | 1.18 | 9.8 | 0.040 | |
| THZ | 111 | 0.96 | 1.110 | 1.25 | 26.2 | 0.065 | 111 | 0.89 | 1.020 | 1.13 | 23.8 | 0.051 | |
| COD3 | 63 | 1.01 | 1.130 | 1.24 | 20.4 | 0.054 | 63 | 0.97 | 1.070 | 1.21 | 22.0 | 0.046 | |
| DIEU | 26 | 1.00 | 1.10 | 1.20 | 18.2 | 26 | 0.97 | 1.08 | 1.20 | 21.2 | |||
| PARR | 14 | 1.01 | 1.100 | 1.17 | 14.7 | 14 | 0.91 | 1.060 | 1.18 | 25.8 | |||
| SAY1 | 3 | 0.95 | 0.990 | 1.05 | 10.0 | 3 | 0.80 | 0.870 | 0.92 | 14.0 | |||
| HERR8 | 12 | 1.02 | 1.085 | 1.18 | 14.5 | 12 | 1.01 | 1.077 | 1.15 | 13.0 | |||
| 27 | 0.90 | 1.010 | 1.12 | 21.8 | 0.067 | 25 | 0.88 | 0.970 | 1.08 | 20.4 | 0.053 | ||
| PDES | 16 | 0.95 | 1.040 | 1.12 | 16.4 | 0.050 | 16 | 0.88 | 0.960 | 1.02 | 14.7 | 0.036 | |
| GAIM | 23 | 0.99 | 1.030 | 1.10 | 10.5 | 0.034 | 22 | 0.94 | 0.990 | 1.05 | 11.1 | 0.032 | |
| RDUBEY | 17 | 0.88 | 1.000 | 1.08 | 20.4 | 0.054 | 17 | 0.84 | 0.951 | 1.05 | 22.2 | 0.051 | |
| VIV1 | 74 | 0.87 | 0.982 | 1.15 | 27.7 | 0.049 | 69 | 0.84 | 0.935 | 1.09 | 25.9 | 0.047 | |
| MIR2A | 24 | 0.91 | 1.006 | 1.08 | 17.1 | 0.050 | 20 | 0.87 | 0.953 | 1.07 | 20.6 | 0.048 | |
| MIR1 | 43 | 0.87 | 1.007 | 1.08 | 21.5 | 0.046 | 44 | 0.84 | 0.967 | 1.10 | 26.8 | 0.054 | |
|
|
|||||||||||||
| M3 | N | Min. | Mean | Max. | V' | σ | N | Min. | Mean | Max. | V' | σ | |
| CLUZEL | 18 | 0.68 | 0.780 | 0.85 | 22.2 | 0.050 | 18 | 0.76 | 0.840 | 0.90 | 16.9 | 0.036 | |
| CHAVR | 15 | 0.66 | 0.740 | 0.86 | 26.3 | 0.055 | 15 | 0.71 | 0.820 | 0.90 | 23.6 | 0.047 | |
| LAUT2 | 20 | 0.71 | 0.780 | 0.90 | 23.6 | 0.048 | 19 | 0.78 | 0.850 | 0.94 | 18.6 | 0.049 | |
| SAULC | 19 | 0.69 | 0.770 | 0.89 | 25.3 | 0.056 | 19 | 0.77 | 0.840 | 0.95 | 20.9 | 0.052 | |
| FORN11 | 7 | 0.72 | 0.774 | 0.82 | 13.0 | 0.034 | 7 | 0.80 | 0.836 | 0.90 | 11.8 | 0.038 | |
| VENINF | 1 | 0.780 | 1 | 0.780 | |||||||||
| BF53 | 5 | 0.92 | 0.950 | 1.02 | 10.3 | 0.040 | 5 | 0.96 | 1.022 | 1.12 | 15.4 | 0.067 | |
| THZ | 87 | 0.67 | 0.790 | 0.90 | 29.3 | 0.044 | 87 | 0.65 | 0.800 | 0.89 | 31.2 | 0.045 | |
| COD3 | 13 | 0.63 | 0.750 | 0.79 | 22.5 | 0.048 | 13 | 0.68 | 0.760 | 0.86 | 23.4 | 0.053 | |
| DIEU | 6 | 0.61 | 0.70 | 0.78 | 24.5 | 6 | 0.66 | 0.75 | 0.84 | 24.0 | |||
| PARR | 2 | 0.79 | 0.830 | 0.87 | 9.6 | 2 | 0.83 | 0.890 | 0.95 | 13.5 | |||
| SAY1 | 1 | 0.670 | 1 | 0.730 | |||||||||
| HERR8 | 11 | 0.74 | 0.824 | 0.89 | 18.4 | 11 | 0.82 | 0.882 | 0.94 | 13.6 | |||
| 9 | 0.68 | 0.750 | 0.82 | 18.7 | 0.046 | 9 | 0.71 | 0.780 | 0.85 | 17.9 | 0.044 | ||
| PDES | 3 | 0.74 | 0.760 | 0.77 | 4.0 | 0.015 | 3 | 0.74 | 0.770 | 0.79 | 6.5 | 0.025 | |
| GAIM | 3 | 0.63 | 0.720 | 0.85 | 29.7 | 2 | 0.82 | 0.860 | 0.89 | 8.2 | |||
| RDUBEY | 4 | 0.64 | 0.738 | 0.80 | 22.2 | 0.068 | 4 | 0.72 | 0.773 | 0.81 | 11.8 | 0.041 | |
| VIV1 | 31 | 0.63 | 0.698 | 0.83 | 27.4 | 0.051 | 29 | 0.66 | 0.733 | 0.81 | 20.4 | 0.041 | |
| MIR2A | 15 | 0.60 | 0.702 | 0.76 | 23.5 | 0.047 | 16 | 0.71 | 0.783 | 0.84 | 16.8 | 0.039 | |
| MIR1 | 37 | 0.63 | 0.700 | 0.77 | 20.0 | 0.035 | 37 | 0.70 | 0.772 | 0.86 | 20.5 | 0.038 | |
A revision of European Plesiosminthus (Rodentia, Dipodidae), and new material from the upper Oligocene of Teruel (Spain)
Plain Language Abstract
Plesiosminthus is an extinct genus of rodents, belonging to the Dipodidae, a family that is currently found across the Northern Hemisphere. The nearest extant relatives of Plesiosminthus are the birch mice (Sicista). The genus Plesiosminthus originated in Asia in the early Oligocene and dispersed into Europe in the late Oligocene. In the early Miocene it became extinct. In this paper we describe two new species and we discuss the European members of the genus. Our material from Spain represents the oldest European occurrence of the genus.
Resumen en Español
Revisión de los Plesiosminthus europeos (Rodentia, Dipodidae), y material nuevo del Oligoceno final de Teruel (España)
En este trabajo revisamos el registro europeo de las especies del género Plesisosminthus y describimos dos nuevas especies. Siguiendo las indicaciones del Código Internacional de Nomenclatura Zoológica, la especie Plesiosminthus promyarion es un nomen dubium y no se debe utilizar. Por primera vez describimos una colección rica del Oligoceno superior (MP 27) de España. Hasta ahora sólo se conocían Plesiosminthus en yacimientos más recientes (MP 28). En Europa se conocen diez especies de Plesiosminthus: nueve del Oligoceno final y una del Mioceno inicial. Para la mayor parte de estas especies no es posible establecer relaciones de parentesco antecesor-descendiente.
Palabras clave: Mammalia, Dipodidae, Oligoceno, Mioceno, nuevas especies.
Traducción: Authors
Résumé en Français
Une révision des Plesiosminthus (Rodentia, Dipodidae) européens et une description de nouveau matériel de l'Oligocène supérieur de Teruel (Espagne)
Le registre fossile européen de Plesiosminthus, un genre de rongeurs, est révisé et deux nouvelles espèces sont décrites. Plesiosminthus promyarion est considéré comme un nomen dubium d'après le Code International de Nomenclature Zoologique et ce nom ne devrait donc plus être utilisé. Jusqu'à présent, ce genre était quasiment inconnu dans les localités plus anciennes que MP28. Pour la première fois, nous décrivons des échantillons conséquents provenant de l'Oligocène supérieur (MP27) d'Espagne. Dix espèces européennes sont connues pour l'instant, une du Miocène inférieur, et les autres de l'Oligocène supérieur. Pour la plupart d'entre elles, il n'est pas possible d'identifier des relations d'ancêtres à descendants.
Mots-clés : Mammalia ; Dipodidae ; Oligocène ; Miocène ; nouvelle espèce
Translator: Antoine Souron
Deutsche Zusammenfassung
Überarbeitung des europäischen Plesiosminthus (Rodentia, Dipodidae) und neues Material aus dem oberen Oligozän von Teruel (Spanien)
Der europäische Nachweis der Nagetier-Gattung Plesiosminthus wird überarbeitet und zwei neue Arten werden beschrieben. Es wurde festgestellt, dass Plesiosminthus promyarion im Sinne der internationalen Regeln der Zoologischen Nomenklatur ein nomen Dubium ist und dass dieser Name nicht länger benutzt werden sollte. Bis jetzt war die Gattung aus Fundstellen die älter als MP28 sind kaum bekannt. Wir beschreiben zum ersten Mal reichhaltige Proben aus dem oberen Oligozän (MP 27) von Spanien. Bis jetzt sind 10 europäische Arten bekannt, eine aus dem unteren Miozän und die anderen aus dem oberen Oligozän. Bei den meisten können keine Vorfahren-Nachfahren-Beziehungen festgestellt werden.
Schlüsselwörter: Mammalia; Dipodidae; Oligozän; Miozän; neue Art
Translator: Eva Gebauer
Arabic

Translator: Ashraf M.T. Elewa
-
-
-
Review: The Princeton Field Guide to Mesozoic Sea Reptiles
 The Princeton Field Guide to Mesozoic Sea Reptiles
The Princeton Field Guide to Mesozoic Sea ReptilesArticle number: 26.1.1R
April 2023


 Poster Winners 2024
Poster Winners 2024