Molecular paleontology as a field engenders much controversy, particularly with respect to the recovery and analyses of truly ancient molecules, i.e. those more than tens of thousands of years old. In part this is due to empirical hypotheses regarding the ultimate durability and survivability of molecules in the fossil record, and in part due to the problems of differentiating between endogenous molecules and exogenous contamination. Molecular and chemical analytical methods are expensive and destructive to rare fossil material, and usually, those fossils most amenable to molecular analyses and most likely to yield positive results are those preserved in an exceptional manner, thus making them too valuable for destructive analyses. The value of molecular paleontology and the techniques and methods it applies, then, is often called into question.
Molecular paleontology, like molecular biology, has come to refer to the recovery, analyses and characterization of DNA. However, the molecular record of an organism is certainly retained in molecules other than DNA, although these other life molecules are variably informative with respect to phylogenies, evolutionary history and other characteristics. Analyses of proteins, carbohydrates, and lipids, as well as their degradation products and products of their interactions with geochemicals, should also be considered aspects of molecular paleontology. Molecular paleontology, then, could be defined as the study of all biomolecules or their degradation products that can be traced to their source and that can shed light on the molecular diagenetic history of an organism.
One assumption that has been pervasive in paleontological thought since the inception of the science is that the organic constituents of an organism, namely soft tissues, cells, or the proteins and nucleic acids which were produced by its living cells, were either destroyed in the process of fossilization (Allison 1990), or rendered uninformative by the diagenetic changes accumulated during geological time (Curry 1990). With advances in the fields of analytical biochemistry, molecular biology, and geochemistry, it is becoming increasingly evident that this is not always the case, and that there may be a wealth of information to be gained through the study of molecular fragments preserved in the fossil record. Examination of such molecules may strengthen the objectivity of the scientific discipline of paleontology, as well as providing an independent means of testing phylogenetic hypotheses.
Molecular paleontology in the modern sense probably began with the report by Abelson (1956) of the recovery of proteinaceous components of fossils. As technology expanded and increased in accuracy, sensitivity, and reliability, new analytical methods began to be applied to fossil material.
In 1974, deJong et al. demonstrated the retention of the antigenic components of proteins preserved within 70 Ma mollusk shells by precipitation reactions with antisera. These results were supported by immunogenic reactivity in other Cretaceous fossil shells (Weiner et al. 1976; Westbroek et al. 1979). Amino acid analyses undertaken by Armstrong et al. (1983) showed the presence of amino acids in a variety of bone samples, and in 1991, Gurley et al. reported the isolation and identification of amino acids in the bony tissues of the sauropod dinosaur Seisomosaurus.
Identification of amino acids within fossil materials does not necessarily imply, however, that those amino acids are derived from original and ancient proteins, as this method does not differentiate between endogenous molecules and those that may have accumulated at any point during diagenesis. Because all organisms on this planet use only the L form of amino acids to build proteins, and these L-amino acids racemize to an equilibrium mixture of D and L isomers after death, chiral analyses of amino acids has been suggested as a means of ruling out the possibility of modern contamination, as a preponderance of L-amino acids may be indicative of an extant or recent origin of protein fragments (Schroeder and Bada 1976). In addition, since each amino acid racemizes to completion at a different rate, it was hypothesized that the degradation of amino acids to a racemic mixture of their D/L isomers may be linked to the age of specimen (Schroeder and Bada 1976; Bada 1985). This latter proposal has been met with some controversy (e.g., Kimber and Griffen 1987), and a method for verifying the endogeneity of proteinaceous material using both racemization analyses and stable isotope geochemistry has been proposed (Macko and Engel 1991).
Further attempts to identify endogenous molecules in fossil materials were undertaken by Lowenstein (1980, 1981, 1985), who demonstrated that chemical extractions of fossil bone were amenable to immunological analyses in the form of solid phase radio-immunoassays. He demonstrated antibody binding to extracts of fossil material from a variety of bone samples, including human, which dated to two million years BP. Based upon his results, he proposed utilizing immunological methods to elucidate phylogenetic relationships of extinct organisms (Lowenstein 1985, 1988). It is now widely accepted that DNA and proteins may be retained in recent fossils or subfossils, although there is much skepticism regarding such preservation in fossils tens of millions of years old.
Early work seeking to identify proteins preserved in fossil material focused on the identification of collagen, because the presence of collagen can be verified by electron microscopy, owing to its unique cross-banded pattern (Van der Rest 1991). However, it was also shown that even preservation at this level of microstructure does not necessarily indicate the presence of endogenous molecules, as collagen-specific amino acids hydroxyproline and hydroxylysine were not identified in samples in which collagen cross-banding could be visualized (Towe and Urbanek 1972).
While demonstrating the presence of amino acids or identifying structural proteins was the goal of early attempts at molecular paleontology, the recognition that some molecules were very durable and that some may have better survival potential than others (e.g., Runnegar 1986) led to the search for other, perhaps more informative proteins, as well as non-proteinaceous material. In addition to collagen (e.g., Baird and Rowley 1990), proteins such as IgG and albumin (Tuross 1989; Cattaneo et al. 1992) have been shown to be preserved in fossil bone, and the vertebrate-specific protein osteocalcin has been identified in both tooth and bone samples (Ulrich et al. 1987) including those of dinosaurs (Muyzer et al. 1992). Hemoglobin, the protein involved in oxygen transport, has also shown potential for preservation in the fossil record, having been identified in association with stone tools (Loy 1983, 1987; Loy and Wood 1989) as well as ancient bone samples (Ascenzi 1985; Smith and Wilson 1990; Cattaneo et al. 1990). Hemoglobin is important both as an indicator of physiology (Dickerson and Geis 1983) and for studies in phylogenetic divergence (Perutz 1983; Nikinmaa 1990; Gorr 1998), and the possibility of its presence in dinosaur bone (Schweitzer et al. 1999) may shed light upon questions of metabolic rates, as well as the relationship of these animals to modern taxa.
Despite these intriguing results, however, the ultimate success of molecular paleontology is viewed by some to be the identification and recovery of DNA sequences from extinct taxa. Of all the biomolecules produced by an animal, DNA contains the most phylogenetic information in its sequences. Data bases now exist that allow comparison of sequences obtained from fossil specimens with those of extant taxa (e.g., Handt et al. 1994; Cooper 1994; Erlich et al. 1991; Pääbo et al. 1989) to test phylogenetic hypotheses (e.g., Felsenstein 1981, 1993; Kumar and Hedges 1998) and to infer evolutionary distance (Lewontin 1989; Hedges et al. 1990, Dolittle et al. 1996). These methods provide the investigator with a means to establish the endogeneity of ancient DNA by placing recovered sequences in correct phylogenetic contexts.
The prevailing scientific opinion has long held that DNA is unstable and easily degraded; therefore its presence in tissue samples much older than a hundred thousand years is highly suspect (Curry 1990; Lindahl 1993). As our understanding of the chemical nature of this molecule increases, it is becoming evident that certain factors act to stabilize DNA, thus significantly increasing its longevity. Desiccation, protection from oxidative damage through rapid burial, and presence of a mineral substrate to which the molecule may adsorb and thus become stabilized, all enhance the preservation potential of DNA (Eglington and Logan 1991; Tuross 1994). More efficient means of extraction (Hoss and Pääbo 1993), coupled with the use of chemical agents to free DNA from complexes of degradation products (Poinar et al. 1998) also increase our chances of success in the identification and recovery of endogenous molecules from the fossil record. The advent of the polymerase chain reaction (PCR) opened the door to the possibility that DNA may indeed be recovered from very old fossils because this reaction makes it possible to amplify small and degraded or altered DNA fragments that perhaps would not be suitable for cloning. However, the sensitivity of PCR creates problems in the analysis of ancient specimens, the most notable of which are the ease with which modern contaminating molecules are amplified and the misleading results due to template damage in ancient samples (Pääbo et al. 1990; DeSalle et al. 1993; Handt et al. 1994).
Mechanisms for the preservation of organic compounds such as DNA or protein over the course of geological time remain to be elucidated. However, it has been proposed that these compounds, in the process of degradation and bond breakage, may co-react to form complex biopolymers which resist further degradation (Curry 1990).
A second mechanism which has been proposed for preservation of biomolecular materials is the stabilization of these molecules through complex interactions with organic breakdown products of the surrounding soils, in particular humic or fulvic acids (Tuross 1994). These associations, while an important factor in the preservation of biomolecules, are also deleterious from an analytical standpoint. Separating out the endogenous components from the rest of the aggregation in order to perform various analyses has proven difficult, although the compound N-phenacylthiazolium bromide (PTB; Poinar et al. 1998) has been demonstrated to be effective in cleaving glycosidic bonds involved in these molecular aggregates, freeing the components of interest. In addition, humic acids fluoresce at the wavelengths of some proteins, amino acids, or nucleic acids (Tuross and Stathoplos 1993), and may therefore interfere with or mask indigenous biomolecular signals. Finally, these breakdown products inhibit the action of some enzymes that may be used to identify organic remains (Tuross 1994), such as digestive enzymes or the polymerase enzymes used in PCR.
Another factor contributing to the preservation of endogenous biomolecules is the early cementation of surrounding or entombing sediments, often facilitated by microbes. This cementation creates a virtually closed system that greatly reduces exogenous degradation processes. Microbes produce extracellular polymeric substances that trap minerals, thus contributing to cementation and their own fossilization as well as the mineralization or "fossilization" of components in the environment. Microbial influences have been invoked in the formation of early diagenetic concretions around biological specimens, which can lead to extraordinary preservation of macro- and microstructure, occasionally including soft tissues (Canfield and Raiswell 1991).
Finally, it is noted in the literature that a primary factor in preserving both proteins and nucleic acids over geological time may be the association of these proteins and/or nucleic acids to a mineral substrate, such as is found in bone (Runnegar 1986; Tuross et al. 1989; Ambler and Daniel 1991; Logan et al. 1991). Adsorbance of biomolecules to minerals may be among the most important of mechanisms involved in biomolecular preservation. Preservation potential is enhanced in biomineralized tissues because there is a component of protein that is encased within the mineral crystals, creating a closed system (Weiner et al. 1989; Sykes et al. 1995).
No doubt, the preservation of biomolecules over the course of geological time is enhanced by a combination of the above mechanisms to varying degrees and, most likely, there are other interactions involved in molecular preservation that have yet to be identified. However, there is little experimental evidence for a temporal limit to preservation enhanced by such mechanisms.
The fossil record is capricious in its preservation. Whereas most fossils are well permineralized, individual specimens can show little evidence of permineralization, which may be an indication of minimal water infiltration. Additionally, surprisingly delicate structures, such as feather barbules or embryonic tissues, can sometimes be seen, and, in fossil Lagerstatten such as the Messel Shale (Schaal and Ziegler 1988), pigment, hair, and individual sarcomeres of muscle fibers have been preserved. Such intricately preserved specimens can reveal additional details which contribute to our understanding of how extinct organisms lived, looked and functioned. For example, the impressions of feathers in the burial sediments surrounding Archaeopteryx led to its placement in the bird lineage, while the presence of feathers and other integumentary structures in exceptionally preserved dinosaurs (e.g., Chen et al. 1998; Quiang et al. 1998; Mayr et al. 2002) not only support the phylogenetic link between dinosaurs and birds (e.g., Gauthier 1986; Sereno 1997), but may also suggest increased metabolic strategies in this group of dinosaurs (Schweitzer and Marshall 2001), consistent with the hypothesis that the origin of birds lies within the Dinosauria. The discovery of oviraptor eggs (Norell et al. 1994) and sauropod eggs (Chiappe et al. 1998) containing delicate embryonic tissues may help to illuminate parental behaviors among dinosaurs, and may shed light on aspects of ontogeny in these taxa. However, little has been done until lately to examine the possibilities of preservation of the molecules that constituted the fossil organisms. This may be due in part to the rarity of appropriate fossil finds, which precludes destructive analyses, and in part to the fact that adaptations of technologies developed for the field of molecular biology have only recently been applied to fossil specimens.
The popularity of the "Jurassic Park" series of books and movies, and the enduring magical fascination dinosaurs holds for the general public, makes inevitable the question "will advances in molecular paleontology ever allow us to resurrect the dinosaurs"? This is a question that paleontologists surely face often since the release of the movie series, and it is to the advantage of students of dinosaur paleontology to understand the issues involved, and to have a clear and concise answer ready when asked.
While no one can predict the future or the directions in which advances in technology will lead us, my answer to the question of dinosaur cloning is a definitive "no". There are several reasons beyond technological problems that lead to this conclusion. First, the successful cloning of a dinosaur requires the recovery of DNA. Proteins, lipids or carbohydrates are insufficient to direct the ontogeny of a living being, as they are simply the indirect or direct products of information stored in the base sequence of DNA. A full complement of DNA is needed to produce a functioning being. In humans, there are more than 108 base pairs of DNA, coding for more than 30,000 genes, all arranged in a specific order and distributed among 46 chromosomes, and each one is necessary to produce a functioning human, orders of magnitude more than the 200–500 bases of endogenous DNA that has been recovered from fossil material. Additionally, if these genes become rearranged, or if a chromosome is lost, or if something else happens to alter the ORDER of base pairs, genes, or chromosomes, it is usually lethal, and almost always severely deleterious, greatly affecting the survivability of the organism.
We have no way of knowing how many genes or chromosomes each dinosaur taxon had, and we cannot deduce the order in which genes were arranged upon individual chromosomes. The order of arrangement is absolutely critical to the development of an organism from a single fertilized cell to a multicellular functioning being.
While the idea of filling in this "missing" genetic information with genes from living, related organisms, as presented in the movie "Jurassic Park", is intriguing, to be successful these genes would have to be almost identical to those of the original dinosaurs, containing the same information and dictating the same functions. We do not possess the information needed to deduce that, and by far the greatest likelihood is that we would end up with a mess of genetic "soup" that would be utterly non-functional.
However, suppose that we could recover, either from the fossil record, or by piecing together the information from living taxa, the total DNA to encode a dinosaur, and that it was arranged in the proper order, upon the exact number of chromosomes. Would we have what we needed then to "grow" a dinosaur? No, it is much more complicated than that, as we would still need an environment in which this genetic information could develop.
In all living animals, development is directly influenced by hormonal cues from the mother. In egg-laying animals, these cues are contained in the yolk and membranes deposited with the embryo. In animals that develop internally, the cues are provided as a continuous flux through the circulatory system, delivered over time to the developing embryo. To "grow" a dinosaur, we would have to take the packet of complete genetic information and insert it into the enucleated egg of a closely related taxon, such as a bird or crocodile, then place that egg within the host animal’s reproductive tract so that it could access the necessary external cues needed to develop. However, the hormonal and environmental cues for development vary greatly within living members of the closest dinosaur relatives. In Crocodilia, gender is determined by the temperature of incubation (Bull 1983; Woodward and Murray 1993). Eggs laid closer to the perimeter of a nest are subjected to colder temperatures, and are usually of a different gender than those incubated within the middle of the nest (Woodward and Murray 1993). In all living birds, on the other hand, sex of offspring is determined genetically, and usually, though not always, the genes determining gender are located on specific chromosomes. We do not know which reproductive strategy the dinosaurs possessed.
Additionally, we do not know the specific hormones
involved, their timing, or the amounts needed to turn on and off the genes of
development. The divergence of the dinosaur–bird lineage from the ancestors of
today’s crocodiles is estimated to have taken place about 230–250 Ma in the
past (Carroll
1988). 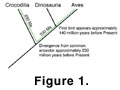 That means that there would be at least 460 million years
of independent evolution between a living crocodile and extant birds, with
dinosaurs diverging at some point along this continuum (see Figure
1). There is
certainly no way of knowing if today’s crocodiles are adequate hosts, at the
molecular level, for the development of a resurrected dinosaur. Similarly, birds
diverged from dinosaurs, it is assumed, at least 150 million years ago. Can we
possibly reconstruct the molecular environments in which a dinosaur embryo
developed?
That means that there would be at least 460 million years
of independent evolution between a living crocodile and extant birds, with
dinosaurs diverging at some point along this continuum (see Figure
1). There is
certainly no way of knowing if today’s crocodiles are adequate hosts, at the
molecular level, for the development of a resurrected dinosaur. Similarly, birds
diverged from dinosaurs, it is assumed, at least 150 million years ago. Can we
possibly reconstruct the molecular environments in which a dinosaur embryo
developed?
But suppose we could overcome these hurdles, and get a viable dinosaur to hatch. What would it eat? The enzymes in its digestive tract would have evolved for specific foodstuffs that in all likelihood no longer exist and have not left any living descendants. If it is a carnivorous dinosaur, could its digestive enzymes break down the components of mammalian tissues? Or would byproducts of digestion be toxic? If it is an herbivore, would today’s plants give it sufficient nutrition, or would it possess the enzymes needed to extract the nutrients from the plant tissues?
These enormous hurdles would have to be dealt with in attempts at resurrecting any extinct animal. They are much easier to address, however, with taxa like the mammoth, having extant relatives, living elephants, that are extremely closely related, and where not much time has elapsed since their divergence and/or extinction. The farther back in time, the more difficult these problems become. Therefore, if one’s goal in the study of molecular paleontology is to resurrect animals that have become extinct, the future of the science is bleak.
Even if the likelihood of building a
"real" Jurassic park is virtually non-existent, there are many
important and interesting questions that can be addressed by applying molecular
techniques to fossil specimens. For appropriately preserved fossil material that
still retains usable molecular information, it may be possible to compare
fragments of molecules with those of close living relatives to estimate the rate
and direction of evolutionary change. For example, if a 200 base-pair fragment
of DNA (e.g., the hemoglobin gene) with 40 informative sites could be
recovered from exceptionally preserved bone tissues of a Velociraptor, it
would be possible to align the dinosaur gene region with the comparable region
of extant crocodiles and birds, and to identify the types of changes between
gene sequences (Figure 2). ![]() In addition, by comparing small regions of genes for
changes, one could infer the closeness of relationships between extinct taxa and
their extant descendants (e.g., Cooper
1994).
In addition, by comparing small regions of genes for
changes, one could infer the closeness of relationships between extinct taxa and
their extant descendants (e.g., Cooper
1994).
Another advantage to studying molecular fragments preserved within fossil tissues would be to date absolutely the timing and direction of genetic changes within taxa, because it would be rooted by the absolute date for the fossil (e.g., van Tuinen and Hedges 2001). This would give us an idea of how long it took genetic changes to accumulate in a lineage, as well as allowing us to infer the number of individual evolutionary events (Lewontin 1989).
The study of remnant molecules in fossils also allows us to understand the processes of molecular diagenesis, or changes that accumulate in the molecule as the result of degradation, modification, and interaction with geochemical residues, and factors within the depositional environment that contribute to the preservation of these same molecules.
All biomolecules break down over time, owing to the action of autolytic enzymes, microbial influences, oxidation, hydrolytic damage, or intra- or intermolecular crosslinking. In addition, DNA molecules can become depurinated or deaminated, or the sugar–phosphate backbone can be cleaved, leaving fragments (Curry 1990). Likewise, proteins can be denatured to primary structure. Once this occurs, original amino acids can convert to others, combine and form cross-links, or lose R-groups completely, leaving any amino acid altered to glycine. Amino acids can also undergo polycondensations, through processes such as Amadori rearrangements or Maillard reactions, leaving insoluble residues containing parts of the original molecules within complexes containing other organic solutions. If one knows the starting molecules, these chemical changes may be able to be elucidated and quantified through the recovery of molecules from fossils. In addition, understanding the type of changes molecules undergo will allow us to predict the chances of molecular recovery of fossils in various environments.
While we will almost certainly never clone a Tyrannosaurus rex, swim with a giant plesiosaur, or breathe new life into the pterosaurs, the field of molecular paleontology has much to offer. Despite the technical problems inherent in dealing with ancient biomolecules and their derivatives, evidence is accumulating that biomolecules, fragments of molecules, or their degradation products can indeed be preserved over geological time scales. Better understanding of the processes of molecular degradation and fossilization, as well as the processes of biomineralization at the molecular level, are shedding light on more efficient means of extracting molecules from fossils (Poinar et al. 1998), which types of molecules may be the best targets for molecular investigations, and on which fossils in which environments may be most appropriate for molecular investigations.
In short, molecular paleontology is a new field, the potential of which is only beginning to be realized. As technologies advance, we will no doubt be able to recover more and more information from the physical remains of animals long extinct. The pursuit of this knowledge is valuable, and will aid in our understanding of evolutionary processes, as well as the processes of fossilization, particularly at the molecular level. Additionally, such studies will clarify our understanding of the stages in the breakdown and modification of molecules over time, thus allowing us to link preserved molecular markers in the fossil record with their source molecules. Finally, understanding molecular diagenesis across geological time scales and recognizing preserved biomarkers from the fossil record may aid in our search for evidence of life on other planets. The search for extraterrestrial life rests on three possibilities, namely, life may never have existed, life may have existed for a short time, then gone extinct, or life may be currently thriving. If the second situation occurs, all that may be left as evidence are resistant molecular markers that are unique to life. We must be able to recognize the range of diagenetic alteration of biomolecules across time on this planet in order to detect them on other planets, where life may have gotten a tenuous start, and then became extinct. Molecular paleontology has much to contribute to the search for life on other planets, in addition to addition to our understanding of the evolution and extinction of life on this one.