RESULTS
 In
the California Bight data 14 samples were available. Half of these samples have
been used as the training set (seven observations). The remaining half
constituted the test set (also seven observations). To make sure that the error
rate is representative of the dataset, the network was run five times, each with
50% random training-set and 50% test-set members (the observations for the
training sets are in italics in Table
1).
In
the California Bight data 14 samples were available. Half of these samples have
been used as the training set (seven observations). The remaining half
constituted the test set (also seven observations). To make sure that the error
rate is representative of the dataset, the network was run five times, each with
50% random training-set and 50% test-set members (the observations for the
training sets are in italics in Table
1).  The
number of network configurations attempted was 600 (20 generations of 30
populations each). Best network configurations for the various partitions are
shown in Table 3. The average
Root-Mean-Square-Error of Prediction (RMSEP) in the test sets was 0.68, implying
that an unknown SST can be predicted from the relative abundances of the five
nannoplankton species used here with a precision of ± 0.68°C (Figure
3). Correlation coefficients between observed and predicted SSTs in the five
test sets range between 0.80 and 0.98 (Figure
4).
The
number of network configurations attempted was 600 (20 generations of 30
populations each). Best network configurations for the various partitions are
shown in Table 3. The average
Root-Mean-Square-Error of Prediction (RMSEP) in the test sets was 0.68, implying
that an unknown SST can be predicted from the relative abundances of the five
nannoplankton species used here with a precision of ± 0.68°C (Figure
3). Correlation coefficients between observed and predicted SSTs in the five
test sets range between 0.80 and 0.98 (Figure
4).
 For
the Mediterranean data the number of samples was 48. We trained the network
using 80% of the samples as the training set (39 observations) and the remaining
20% as the test set (nine observations). This subdivision of the training and
test sets was performed randomly by the program. Again, five different random
subdivisions of the original data set were run to generate an error estimate.
Also in this case, the number of network configurations attempted was 600 (20
generations of 30 populations each).
For
the Mediterranean data the number of samples was 48. We trained the network
using 80% of the samples as the training set (39 observations) and the remaining
20% as the test set (nine observations). This subdivision of the training and
test sets was performed randomly by the program. Again, five different random
subdivisions of the original data set were run to generate an error estimate.
Also in this case, the number of network configurations attempted was 600 (20
generations of 30 populations each).  The
configurations of the best networks are provided in Table
3. The average RMSEP in the test sets was 0.64, implying that an
unknown oxygen isotope value can be predicted from the relative abundances of
the selected nannofossil species in this core with a precision of ± 0.64
The
configurations of the best networks are provided in Table
3. The average RMSEP in the test sets was 0.64, implying that an
unknown oxygen isotope value can be predicted from the relative abundances of
the selected nannofossil species in this core with a precision of ± 0.64 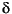 18O
18O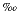 PDB (Figure 5). Correlation
coefficients between observed and predicted isotope data in the five test-sets
range between 0.64 and 0.96 with an average value of 0.88 (Figure
6).
PDB (Figure 5). Correlation
coefficients between observed and predicted isotope data in the five test-sets
range between 0.64 and 0.96 with an average value of 0.88 (Figure
6).
 In
the California Bight data 14 samples were available. Half of these samples have
been used as the training set (seven observations). The remaining half
constituted the test set (also seven observations). To make sure that the error
rate is representative of the dataset, the network was run five times, each with
50% random training-set and 50% test-set members (the observations for the
training sets are in italics in Table
1).
In
the California Bight data 14 samples were available. Half of these samples have
been used as the training set (seven observations). The remaining half
constituted the test set (also seven observations). To make sure that the error
rate is representative of the dataset, the network was run five times, each with
50% random training-set and 50% test-set members (the observations for the
training sets are in italics in Table
1).  The
number of network configurations attempted was 600 (20 generations of 30
populations each). Best network configurations for the various partitions are
shown in Table 3. The average
Root-Mean-Square-Error of Prediction (RMSEP) in the test sets was 0.68, implying
that an unknown SST can be predicted from the relative abundances of the five
nannoplankton species used here with a precision of ± 0.68°C (Figure
3). Correlation coefficients between observed and predicted SSTs in the five
test sets range between 0.80 and 0.98 (Figure
4).
The
number of network configurations attempted was 600 (20 generations of 30
populations each). Best network configurations for the various partitions are
shown in Table 3. The average
Root-Mean-Square-Error of Prediction (RMSEP) in the test sets was 0.68, implying
that an unknown SST can be predicted from the relative abundances of the five
nannoplankton species used here with a precision of ± 0.68°C (Figure
3). Correlation coefficients between observed and predicted SSTs in the five
test sets range between 0.80 and 0.98 (Figure
4).
