 Quantifying the living fossil concept
Quantifying the living fossil concept
Article number: 21.1.14A
https://doi.org/10.26879/750
Copyright Palaeontological Association, April 2018
Author biographies
Plain-language and multi-lingual abstracts
PDF version
Submission: 2 December 2016. Acceptance: 10 April 2018
{flike id=2194}
ABSTRACT
“Living fossil” is a contentious label, often used to identify clades that have experienced particularly little evolutionary change. Many of the problems associated with the term are due to a lack of a clear definition. To date, most work on the phenomenon has been primarily qualitative, leading to a list of living fossils each selected for different sets of reasons. This non-uniformity in living fossil identification makes the ubiquity, clarity and potential causes of the phenomenon difficult to assess. An alternative approach is to use a quantitative metric that matches the most common interpretations of “living fossil” to generate a less subjective listing. Here, we present the Evolutionary Performance Index (EPI); this metric is calculable across the entire tree of life and allows for fair comparisons between taxonomic groups. With this index, we calculated the performance scores for over 24,000 clades within Metazoa and Embryophyta. Many well-known living fossils featured among the lowest performing clades, e.g., coelacanths, gingko, tuatara as well as groups that have previously been overlooked. By grounding the definition in a strictly quantitative framework, future researchers will be better able to test the causes and relevance of the phenomenon.
Dominic J. Bennett. Department of Earth Sciences and Engineering, Imperial College London, London UK. Institute of Zoology, Zoological Society of London, London UK. dominic.john.bennett@gmail.com
Mark D. Sutton. Department of Earth Sciences and Engineering, Imperial College London, London UK. m.sutton@imperial.ac.uk
Samuel T. Turvey. Institute of Zoology, Zoological Society of London, London UK. samuel.turvey@ioz.ac.uk
Keywords: Living fossil; evolutionary distinctness; evolutionary performance; evolutionary performance index
Final citation: Bennett, Dominic J., Sutton, Mark D., and Turvey, Samuel T. 2018. Quantifying the living fossil concept. Palaeontologia Electronica 21.1.14A 1-25. https://doi.org/10.26879/750
palaeo-electronica.org/content/2018/2194-quantifying-the-living-fossil
Copyright: April 2018 Palaeontological Association.
This is an open access article distributed under the terms of Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0), which permits users to copy and redistribute the material in any medium or format, provided it is not used for commercial purposes and the original author and source are credited, with indications if any changes are made. creativecommons.org/licenses/by-nc-sa/4.0/
INTRODUCTION
Darwin (1859) coined the term “living fossil” to mean a species or group of species that has remained so little changed that it provides an insight into earlier, now extinct, forms of life. Despite Darwin’s admission that the label was “fanciful”, the term has persisted. Classic examples of living fossils are horseshoe crabs (family Limulidae), tuatara (Sphenodon) and the ginkgo (Ginkgo biloba). These groups first appeared in the Ordovician, the Triassic, and the Permian, respectively, they have changed little from their fossil ancestors and all have few surviving species (Royer et al., 2003; Hay et al., 2008; Rudkin et al., 2008). Recently, the term has gained increased attention due to genomic studies indicating that “living fossil” taxa such as coelacanths (Latimeria) and elephant sharks (Callorhinchus) have, respectively, experienced lower rates of genomic rearrangement (Smith et al., 2012; Amemiya et al., 2013) and protein sequence evolution (Venkatesh et al., 2014) than other vertebrate genomes. Additionally, large-scale studies of diversification rates for multiple taxonomic groups have discovered certain ‘living-fossil-like’ lineages have experienced lower rates of speciation and extinction (Alfaro et al., 2009; Near et al., 2014).
The living fossil concept is, however, controversial and viewed unfavourably by many evolutionary biologists and palaeontologists. Much of the difficulty surrounding the term, however, stems from its multiple and often vague definitions, which causes different authors to classify different sets of organisms as living fossils (Schopf, 1984). Some authors place increased focus on living fossils constituting “evolutionary relicts” (Nagalingum et al., 2011), others focus on “little change through time” (Eldredge, 1984; Fisher, 1990), and others still interpret the term to mean Lazarus taxa (Smith, 1939). More recently, there have been efforts to reject the living fossil term entirely as it is thought to recall Haeckel’s scala naturae and is a product of bad ‘tree-thinking’ (Casane and Laurenti, 2013; Grandcolas et al., 2014; Minelli and Baedke, 2014). It is argued that the term living fossil is a form of ‘progressivist’ language that can promote a false interpretation of evolution where life is organised into ‘higher’ and ‘lower’ ranks (Rigato and Minelli, 2013). Under a progressivist scheme, these ‘lower’ ranked organisms are then able to subvert post-Darwinian evolutionary thinking by remaining unchanged for millions of years - an impossibility even in the hypothetical circumstance of an absence of selective pressure (Casane and Laurenti, 2013).
Additionally, doubts over the concept are compounded all the more by new evidence. Some groups that were once thought to be species-poor have since been found to be species-rich (Monoplacophora [Kano et al., 2012] and Notostraca [Mathers et al., 2013]); others have been shown to have more genetic diversity than previously thought (Limulidae [Obst et al., 2012]); some show great morphological diversity in the past (Crocodilia [Buckley et al., 2000], Sphenodon [Meloro and Jones, 2012] and Latimeria [Casane and Laurenti, 2013]), and other groups that were thought to consist of ancient lineages have, in fact, experienced most of their diversification relatively recently (Cycadophyta [Nagalingum et al., 2011]).
Most work on the living fossil concept has hitherto been restricted to single groups, and has focused on describing the nature and magnitude of evolutionary changes seen in these groups within the fossil record (e.g., Eldredge, 1984). An alternative approach is a pan-group analysis, which could highlight and resolve inconsistencies around current usage of the term “living fossil”, to produce a more precise and usable definition that maintains some congruence with common usage. A key element of such an analysis would be a quantitative scale of “living-fossil-ness”, combining different aspects of the concept into a single index that could be used to determine which groups may be considered living fossils in a more objective manner. The availability of such a metric would enable a more rigorous analysis of the degree to which living fossils are a phenomenon worthy of study. Such a measure would have to be applicable across the tree of life, be applicable to clades as well as species; be readily measurable; and allow fair inter-group comparisons. No explicit metric of living-fossil-ness has yet been proposed, although evolutionary distinctness (or ED) has been suggested as a proxy (Isaac et al., 2007; Cavin and Kemp, 2011). ED is applicable across the tree of life, allows inter-group comparisons, and captures the element of phylogenetic isolation and lineage antiquity that is common to most interpretations of the living fossil concept. ED, however, is not measurable for clades above the species-level, meaning that many candidate living fossil groups (e.g., coelacanths, monotremes) can only be considered on the basis of their individual species. It also requires a fully resolved, time-calibrated phylogenetic tree of the entire taxonomic group of interest, a substantial impracticality for any metric intending to identify living fossils across all life. Finally, it does not take into account other elements frequently incorporated into the living fossil concept such as morphological or ecological change (Fisher, 1990).
We here propose a new metric to address these shortcomings, and demonstrate its fit to qualitative concepts of “living fossil-ness” in a range of groups. We surveyed the literature to find the most common ways that the term “living fossil” is used, and broke these down into measurable elements: age of the clade (in millions of years), relative success in terms of numbers of species, and relative number of changes since the clade first appeared. We combined these three measurable elements into a single equation to produce an evolutionary performance index (EPI), and calculated EPI values across all metazoans and plants to generate a ranked list of the most living fossil-like clades. Clades that score low on this index are considered “living fossil-like”, while those that score high are not (i.e., have likely experienced recent adaptive radiations). We also introduce pEPI, a proxy for EPI for use where number of changes cannot be calculated. Our indices allow not simply a measure of living-fossil-ness for any given clade, but a breakdown of which elements of the concept (age, relative success, number of changes) are responsible for any particularly low or high value.
MATERIALS AND METHODS
Literature Survey
Using “living fossil” as a keyword, we searched for all literature, scientific articles and books that proposed a definition of the living fossil. We disregarded all sources that used a “progressivist” or a Lazarus taxon interpretation of the term. In cases where a definition statement could not be found, a definition was inferred from the text as a whole. In total we discovered 56 sources from which an explicit definition could be determined (see Appendix 1 for a list of references and Appendix 2 for example of interpretation of text). We identified eight recurring themes from our survey: existing for a long time (1), morphologically conserved (2), some alternative form of conservatism (3), having “primitive” features (4), phylogenetically/evolutionarily distinct (5), a survivor of a once large clade (6), geographically isolated (7) and having a generalist niche (8).
We disregarded themes 7 and 8 as we considered these to be potential explanatory or emergent characteristics of living fossils, rather than defining features. Equally, there is disagreement on the validity of these themes. For example, we might consider living fossils to be generalists because highly ecologically specialised species are more likely to become extinct, as they have more restrictive dietary or habitat requirements, and so are more vulnerable to loss of key food resources or habitats, and thus extinction (Eldredge, 1979; Stanley, 1998). On the other hand, generalist species tend to have broad distributions facilitating allopatric speciation, therefore making the lineage less like a living fossil (Jackson, 1974). Also, although it is often asserted that life-history specialisation should increase a lineage’s risk of extinction, it cannot be determined from the outset. For example, Cieslak et al. (2014), demonstrated that cave beetles have in fact experienced multiple radiations since becoming specialised to a subterranean environment.
Furthermore, there are many examples of clades considered living fossils that are not geographically isolated and generalist in niche (e.g., crocodiles, sharks, moss, horseshoe crabs); equally there are many clades that exhibit these features and are not considered living fossils (recently diverged island species, highly successful generalist clades like rodents and grasses).
Additionally, there is disagreement between authors on whether a living fossil should solely be an unchanging species (e.g., Schopf 1984) or can be a higher-level taxon that may experience low rates of speciation (e.g., Yoshida 2002). For the purposes of this study, we opted to assess any monophyletic group (species or not) as a candidate for a living fossil.
We combined the remaining six themes into three quantifiable variables: “success”, “time” and “change”. These were in turn combined into an “Evolutionary Performance Index”, based on current success and levels of change relative to a clade’s sister (see below for more details).
Success
This variable captures themes 5 and 6. It is calculated simply as the number of extant species. Lineages that have low species counts will be more likely to be evolutionary distinct and be survivors of once large clades. We used the NCBI taxonomy (Federhen, 2012) to estimate success and other variables across all metazoans and plants. NCBI taxonomy was selected over other online taxonomies (e.g., ITIS, 2016) because, as a repository for sequence data, we deemed its names to be the most likely to have associated time-estimates. Furthermore, all clades within the database aim to be monophyletic, and classifications are regularly updated according to the latest studies in the systematic literature (Federhen, 2012). To aid interpretation of results, all recovered clades were assigned to one of nine large, well-known groupings: Aves, Mammalia, Actinopterians, Lepidosauria, Amphibia, other vertebrates, Arthropoda, other metazoans and Embryophyta. We downloaded all relevant taxonomies from NCBI and counted the number of descendent species for every clade. Many names for biological entities in the taxonomy are not necessarily recognised extant species or groups. We, therefore, excluded all “unclassified”, “unassigned”, “unvouchered”, “extinct” and “environmental” entries as well as next-generation sequencer identified species. We ignored this latter group as we could not be certain of the species status. We also identified each sister clade, and calculated the contrasted number of descendants by dividing the clade’s number of descendants by its sister’s. In cases where there was more than one possible sister, i.e., a polytomous node, we selected the sister with the greatest number of descendants in order to tend towards the lowest possible EPI scores.
Time
This variable captures themes 1 and 5 and is calculated as the time, in millions of years, since the lineage diverged. We used two approaches for estimating time. First, we sourced large-scale time-calibrated phylogenetic trees for two major vertebrate groups, mammals (Bininda-Emonds et al., 2007) and birds (Jetz et al., 2012; Birdtree, 2016). Because the source data for the bird tree is composed of two posterior distributions generated from different avian taxonomies, we selected a random set of 100 trees from the distribution based on the Hackett et al. (2008) taxonomy. For every clade in these phylogenetic trees we estimated the clade’s age, determined as the time since the lineage diverged from its closest living sister, using the R-package treeman (Bennett et al., 2017). Common clades between those of NCBI and those found in the phylogenetic trees were identified using name matching of descendants. The second approach used TimeTree (Hedges et al., 2006, 2015; Kumar and Hedges, 2011), an online database of divergence times. This database hosts a list of estimated divergence times across all life as reported in the scientific literature, including all published estimates for the divergence date for two given taxa and a weighted average measure of divergence based on these estimates. Although direct divergence times observed from the fossil record would be better than times inferred from sequence differences, the largest repository of these data (fossilcalibrations.org) is not yet available for automated searching and does not have as many entries. Equally, the data recovered from TimeTree is not simply the average from the estimated divergences for a given split; this repository implements a method that relies on the structure of the tree of life in order to up-weight more reliable time estimates (‘hierarchical average linkage’, Hedges et al., 2015). A demonstration that TimeTree and (manually extracted) fossil calibrations data produce similar results is provided in Appendix 3 fora limited number of clades. We sought the age of a given clade by performing TimeTree searches of the clade against all possible sister clades. In order to estimate the lowest possible EPI values we then selected the highest reported divergence date among all sisters. To reduce computational time and to limit the dependence of our results on averaged estimates of divergence, we conservatively restricted our TimeTree searches only to clades we considered more likely to be living fossils, identified as clades whose parent clade contained more than 500 species and which had a contrasted number of descendants of less than 0.1. This restriction only prevents the unnecessary searching for high EPI clades because parental clades containing fewer than 500 species are likely to have only existed for relatively short periods of time. Additionally,10 times fewer descendants than your sister is a common score even for clades not considered living fossils. Using this cut-off, for example, rabbits fall under consideration (e.g., Oryctolagus [1 sp.] vs. Lepus [32 spp.]: 0.03125).
Change
This variable captures themes 2, 3 and 4 and is calculated as the number of changes (morphological and ecological) that have occurred in the clade’s lineage since its initial divergence. We estimated change from datasets of morphological, ecological and life history traits using ancestral character reconstruction. This required phylogenetic trees in order for the ancestral characters be estimated, and, therefore, we only sourced datasets for the major taxonomic groups for which we had trees and large datasets of characters (mammals and birds). For all other clades, we calculated only pEPI values, which do not require a measure of change (see below). For both mammals and birds, we sourced one morphological character dataset (Livezey and Zusi, 2007; O’Leary et al., 2013) and one ecological/life history dataset (Lislevand et al., 2007; Jones et al., 2009). The names found within the datasets were matched to tips in the phylogenetic trees using character matching. The combined datasets represented 4,572 characters and 4,510 species for mammals, and 2,988 characters and 3,509 species for birds. For character reconstruction we used the R-package ape (Paradis et al., 2004) to implement maximum parsimony reconstruction (Hanazawa et al., 1995). Although it is likely that model-based approaches, particularly using Bayesian methods (e.g., O’Reilly et al., 2016), are superior to parsimony in a phylogenetics context, we chose not to perform ancestral character reconstruction with any model. For the two most common models for character change, Brownian motion or 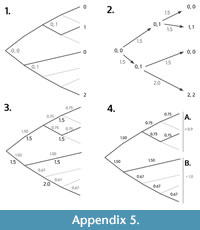 Ornstein-Uhlenbeck, doubts exist over their accuracy (Thomas et al., 2014), there is evidence that they may be prone to undetectable trends (Webster and Purvis, 2002), and they do not always outperform parsimony (Royer-Carenzi et al., 2013). We, therefore, used a parsimony approach when estimating ancestral characters. Continuous traits were made discrete by binning into 10 equally spaced categories. We used the mammalian super tree (Bininda-Edmonds et al., 2007) and a consensus bird tree (Birdtree, 2016) to perform ancestral character reconstruction for all characters. Change scores were calculated for every branch in a tree based on differences between the ranges of possible states of the nodes of the tree. Score values of one indicate no possibility of change, less than two the possibility of change, and more than two that multiple changes must have occurred. Mean change scores per character were calculated for each clade in a tree using the scores of all descendent branches. Sister contrast values were then calculated for each character by dividing the clade mean by the sister mean. An overall change score from these sister contrasted means per character was then determined using a weighted mean to control for non-independence and number of states per character. No score was calculated for any clade that had fewer than four characters represented. (See Appendix 4 and Appendix 5 for more details.)
Ornstein-Uhlenbeck, doubts exist over their accuracy (Thomas et al., 2014), there is evidence that they may be prone to undetectable trends (Webster and Purvis, 2002), and they do not always outperform parsimony (Royer-Carenzi et al., 2013). We, therefore, used a parsimony approach when estimating ancestral characters. Continuous traits were made discrete by binning into 10 equally spaced categories. We used the mammalian super tree (Bininda-Edmonds et al., 2007) and a consensus bird tree (Birdtree, 2016) to perform ancestral character reconstruction for all characters. Change scores were calculated for every branch in a tree based on differences between the ranges of possible states of the nodes of the tree. Score values of one indicate no possibility of change, less than two the possibility of change, and more than two that multiple changes must have occurred. Mean change scores per character were calculated for each clade in a tree using the scores of all descendent branches. Sister contrast values were then calculated for each character by dividing the clade mean by the sister mean. An overall change score from these sister contrasted means per character was then determined using a weighted mean to control for non-independence and number of states per character. No score was calculated for any clade that had fewer than four characters represented. (See Appendix 4 and Appendix 5 for more details.)
Calculation of Indices
EPI (Evolutionary Performance Index) combines the three variables of success, time and change into a scalar. Because performance is relative, EPI uses values of success and change measured as “sister contrasts”, i.e., relative to the equivalent values of the sister clade, rather than raw values. Sister clades are appropriate benchmarks of performance; a clade and its sister share the same evolutionary trajectory until the point of divergence - as originally suggested by Vrba (1984). The use of sister contrasts allows EPI comparisons across taxonomic groups, and also has the benefit of forcing the values of success and change onto the same scale (> 0, skewed distribution). The EPI of a clade can, therefore, be thought of as the amount of evolutionary “activity”, measured as the number of species and changes relative to its sister, over the amount of time a clade has existed. Because it can be difficult to source the data required to measure change, we present two indices, EPI (eq. 1) where all three variables can be estimated, and “proximate” EPI or pEPI (Eq. 2) where only time and success can be estimated:
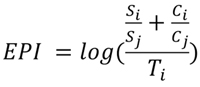 |
(1) |
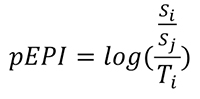 |
(2) |
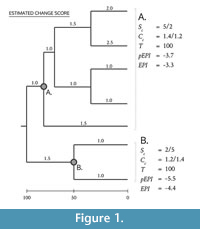 These define the performance for any clade of interest (i) relative to its sister (j). S, C and T refer to the estimated values for success, change and time, respectively, (see Figure 1). Because the resulting division of success and change over time produces a heavily skewed distribution, the logarithm of this ratio is used to generate a normal distribution.
These define the performance for any clade of interest (i) relative to its sister (j). S, C and T refer to the estimated values for success, change and time, respectively, (see Figure 1). Because the resulting division of success and change over time produces a heavily skewed distribution, the logarithm of this ratio is used to generate a normal distribution.
Comparing the Fit of ED, EPI and pEPI to Qualitative Assessments of “Living Fossil-ness”
All three metrics were calculated for every clade where data permitted. For all clades where an index was calculated, we then searched for the clade’s corresponding Wikipedia article (Wikipedia, 2016) and generated presence/absence data on whether the phrase “living fossil(s)” appeared. Whereas this measure is crude, it represents a practical and relatively unbiased assessment of the popular applicability of the term to a clade. We ran generalised linear models with binomial error distributions (Hastie and Pregibon, 1992) to determine whether the three indices correspond to this measure of popular usage.
R (3.2.4) was used to run all analyses. The pipeline used to generate all results is available via the main author’s GitHub account (Bennett, 2016).
RESULTS
In total we calculated pEPI scores for 21,740 clades, with the majority of values being calculated for mammals and birds through the use of time-calibrated phylogenetic trees (10,136 Aves, 5,774 Mammalia, 280 Actinopterians, 103 Lepidosauria, 93 Amphibia, 18 other vertebrates, 2,049 Arthropoda, 296 other metazoans, and 2,992 Embryophyta). The majority of values were negative (pEPI quantiles: -16.3-0%, -6.5-25%, -4.7-50%, -3.3-75%, 3.5-100%). Many of the lowest scoring clades have been considered “living fossils” by previous authors, Trichoplax (-16.2), coelacanths (-15.6), lancelets (-14.9) and limulids (-13.6). For EPI, scores were calculated for 2,433 and 3,012 clades for mammals and birds, respectively. The resulting range of values was less negative compared to pEPI (EPI quantiles: -5.1-0%, -2.8-25%, -2.0-50%, -1.2-75%, 3.7-100%). Again, taxa regularly considered to represent living fossils had the lowest scores, monotremes (-5.4), hoatzin (-4.6), marsupials (-4.9) and palaeognaths (-4.7). The pEPI of Homo sapiens (-2.2) was near to the median value, but for apes overall the score was relatively low for both pEPI (-5.1) and EPI (-3.4). (See Table 1 and Table 2, see Appendix 6 for statistics calculated for all 21,740 clades and Appendix 7 for estimates of contrasted change.)
Many taxonomic groups had lower than expected numbers of species in the NCBI database. Embryophytes, metazoans and particularly arthropods were under-sampled when compared to estimated numbers of species for these groups (see Table 3 for sampled and expected species counts).
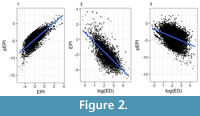 For clades that had both EPI and pEPI scores, there was a strong correlation between the two indices (Pearson’s R = 0.77, Spearman’s R = 0.75; see Figure 2.1). We also found a positive correlation between the change variable and pEPI (Spearman’s R = 0.45). Additionally, both indices showed relatively strong correlations with ED (Pearson’s R = -0.41 and -0.72 for pEPI and EPI respectively against log ED; Figure 2.2-2.3).
For clades that had both EPI and pEPI scores, there was a strong correlation between the two indices (Pearson’s R = 0.77, Spearman’s R = 0.75; see Figure 2.1). We also found a positive correlation between the change variable and pEPI (Spearman’s R = 0.45). Additionally, both indices showed relatively strong correlations with ED (Pearson’s R = -0.41 and -0.72 for pEPI and EPI respectively against log ED; Figure 2.2-2.3).
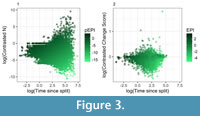
Each variable experienced different levels of dispersion, with success having the greatest coefficients of variation (39.5, 1.2 and 0.2 for success, time and change, respectively). Despite these great differences, all of the variables had an impact on the overall scores (Spearman’s R 0.78, and -0.68 for success and time against pEPI and Spearman’s R 0.21, 0.25 and -0.95 for success, change and time, respectively for EPI). Both indices, also, showed an increasing range of variables towards higher values of time (Figure 3).
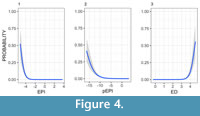
 In total we sourced 13,898 Wikipedia articles for all clades where we had calculated indices. Of these articles, 62 contained the phrase “living fossil”. EPI and pEPI both showed that the lower the value, the more likely a clade’s article is to include the term. Within the bottom 0-25% quantile of EPI and pEPI clades, 23 and 40 included “living fossil” in their articles, whereas there were only 2 and 19, respectively in total in the subsequent three quantiles (25-100%). For ED the pattern was as expected given that higher values indicate greater distinctness: 31 mentions in the 75-100%, and 4 in the 0-75%. We did not identify any of these indices to be significantly better; none of the binomial models for each of the indices differed significantly when explaining the 62 instances of the phrase (residual deviances of each metric modelled against “living fossil” mentions: -1 - 168.4, ED - 115.4, pEPI - 120.8, EPI - 102.9) (see Figure 4).
In total we sourced 13,898 Wikipedia articles for all clades where we had calculated indices. Of these articles, 62 contained the phrase “living fossil”. EPI and pEPI both showed that the lower the value, the more likely a clade’s article is to include the term. Within the bottom 0-25% quantile of EPI and pEPI clades, 23 and 40 included “living fossil” in their articles, whereas there were only 2 and 19, respectively in total in the subsequent three quantiles (25-100%). For ED the pattern was as expected given that higher values indicate greater distinctness: 31 mentions in the 75-100%, and 4 in the 0-75%. We did not identify any of these indices to be significantly better; none of the binomial models for each of the indices differed significantly when explaining the 62 instances of the phrase (residual deviances of each metric modelled against “living fossil” mentions: -1 - 168.4, ED - 115.4, pEPI - 120.8, EPI - 102.9) (see Figure 4).
DISCUSSION
 We have developed practical metrics for determining the extent to which clades can be associated with the living fossil concept. We have demonstrated that these indices, EPI and pEPI, correspond well to the popular conception of the living fossil by showing that low-scoring clades are much more likely to be described as living fossils in their Wikipedia articles. Furthermore, we have demonstrated that the more easily calculated pEPI correlates well with EPI. Many well-known living fossil clades appeared in the top rankings for pEPI, including, among others, coelacanths (Cavin and Guinot, 2014), tadpole shrimps (Mathers et al., 2013), lancelets (Garcia-Fernàndez and Benito-Gutiérrez, 2009), lungfishes (Cavin and Kemp, 2011), limulids (Kin and Błazejowski, 2014), tuatara (Hay et al., 2008, Herrera-Flores et al., 2017), crocodiles (Buckley et al., 2000) and the ginkgo (Royer et al., 2003). By better describing what constitutes a living fossil it will be easier to resolve conflicts over its meaning, and investigate any underlying causes that may lead to the condition.
We have developed practical metrics for determining the extent to which clades can be associated with the living fossil concept. We have demonstrated that these indices, EPI and pEPI, correspond well to the popular conception of the living fossil by showing that low-scoring clades are much more likely to be described as living fossils in their Wikipedia articles. Furthermore, we have demonstrated that the more easily calculated pEPI correlates well with EPI. Many well-known living fossil clades appeared in the top rankings for pEPI, including, among others, coelacanths (Cavin and Guinot, 2014), tadpole shrimps (Mathers et al., 2013), lancelets (Garcia-Fernàndez and Benito-Gutiérrez, 2009), lungfishes (Cavin and Kemp, 2011), limulids (Kin and Błazejowski, 2014), tuatara (Hay et al., 2008, Herrera-Flores et al., 2017), crocodiles (Buckley et al., 2000) and the ginkgo (Royer et al., 2003). By better describing what constitutes a living fossil it will be easier to resolve conflicts over its meaning, and investigate any underlying causes that may lead to the condition.
Overlooked Living Fossils
We identified many clades that have not commonly been considered living fossils, but which have low scores in our indices. These clades are mainly microscopic metazoans that split early in the evolution of eumetazoa. The top three living fossils according to pEPI were the recently discovered Limnognathia maerski, whose likely closest relatives are the rotifers (Kristensen, 2002) from which it split 662 m.y.a.; the amoeboid placozoa (Trichoplax), which is possibly an early diverging sister to all diploblasts (Syed and Schierwater, 2002; Voigt et al., 2004; Schierwater et al., 2009); and the worm-like mesozoans, which are potentially sister to all Lophotrochozoa (Suzuki et al., 2010). Although their exact phylogenetic positions and species counts have not yet been fully resolved, their ages are in the 100s of m.y. and their species counts are below 100; they fit most people’s conception of a living fossil.
We also identified low scoring clades among larger bodied metazoans that have been often overlooked for living fossil status. For example, the mountain beaver (Aplodontia rufa) is sister to all rodents and had one of the lowest scores among all mammals; the hoatzin (Opisthocomus hoazin) is an early diverging neognath had the lowest score among birds and the dwarf pencil fish (Lepidogalaxias salamandroides) was the lowest scoring actinopterian. These animals are not regularly cited as living fossils in the most well-known literature that discuss the phenomenon (e.g., Schopf, 1984; Eldredge and Stanley, 1984; Fisher, 1990), yet our analysis shows they are just as good candidates for the term as many oft-cited examples. By including previously overlooked groups such as these, future work on the common causes of the living fossil phenomenon will be able to sample a broader set of clades.
Taxonomic Hierarchy
A few clades that have been described as living fossils that did not have low scores belonged to larger clades that did. For example, although the term is commonly applied to the cycads, we found this group to have a mid-ranging pEPI score. Nagalingum et al. (2011) argued against cycads being living fossils due to recent radiations within the group, but in our analysis their score is modest as they perform comparably to their sister taxa, which include other “low performing” gymnosperm clades such as conifers, gnetophytes, and Ginkgo. However, the parent clade of these gymnosperm clades, Acrogymnospermae, has a very low pEPI score (in the lowest 2% of plants and metazoans). We hence contend that acrogymnosperms as a whole, rather than the cycads in particular, are good candidates to receive the living fossil label. Equally, although “sharks” (Elasmobranchii, a grouping that includes rays) are commonly described as living fossils, they had a high pEPI score due to their relatively better performance compared to their sister lineage (Holocephali - Chimaeriformes). Again the living fossil label for “sharks” is not so much inappropriate as misplaced in the hierarchy; we found the parent group of both these groups, Chondrichthyes, to have a particularly low pEPI score due to it being sister to all bony fish and tetrapods (Euteleostomi). The Hula painted frog (Latonia nigriventer) provides another example; it was labelled a living fossil when first described (Biton et al., 2013), but, as a species, we did not find it among the top living fossils within Amphibia. Instead, the family (Alytidae, painted frogs) was among the top ten.
These examples demonstrate the benefit of our larger scale approach based around monophyletic clades as it allows us to better pinpoint the full extent of the region of the tree of life that has experienced evolutionary stagnation. Indeed, this transfer of living fossil status to higher-level clades may explain some inconsistencies in our Wikipedia analysis. For example, Anispotera, Macroscelididae and Solenodon paradoxus are all described as living fossils in their articles but have unexceptional EPI/pEPI scores, yet their parents (Palaeoptera, Afrotheria and Solenondon) all have low scores, without necessarily being described as living fossils themselves in Wikipedia.
Data Limits
We argue that pEPI and EPI are less prone to sampling bias compared to ED because they only require the clade and its sister to have equal levels of sampling rather than a complete species-level phylogenetic tree. Despite this, it was evident that the relative sampling of the different clades impacted the scores. Arthropods had less than three percent of their expected number of species accounted for in the NCBI taxonomy. According to the pEPI ranking, among the top 10 clades there were three lepidopteran species: Agathiphaga queenslandensis, Heterobathmia pseuderiocrania and Andesiana lamellata. We would not expect these Lepidoptera to have lower scores than other low diversity, early-branching (“basal”) arthropod clades - such as the Diplura, Protura, Zygentoma or Limulidae. Instead, it is likely that the unexpected low scores for these species are due to the order of magnitude more DNA barcode records for Lepidoptera than for other arthropods (BOLD, 2016). For example, although it is well known that Coleoptera is the most species-rich insect order, in the NCBI database it is less sampled (26,240 spp.) than Lepidoptera (37,934 spp.). Low sampling within a group is more likely to affect larger clades than smaller ones because researchers tend to sample a range of diverse groups, e.g., for phylogenetic analysis. With the amelioration of taxonomic sampling of DNA databases, we would, therefore, expect low-scoring clades within the under-sampled plants and arthropods to have still lower scores.
Variable Dominance
All the variables had an impact on the resulting EPI and pEPI values. In the case of EPI, however, the score was largely determined by the time variable. This led to fewer single species clades ranking at the bottom. This was in part due to the stabilising impact of the change variable when combined with the success variable. For example, the Palaeognathae that ranked among the bottom 10 birds for pEPI, ranked as having the lowest EPI score. Although its success (0.06) and change (1.01) variables were not as low as for other bird groups (e.g., 0.0006 and 0.849 for Menuridae, 0.0003 and 1.00 for Upupiformes), when added they were closer to the values of other groups - making the time variable the deciding factor. As a consequence, fewer single species clades, which tend to be younger, ranked among the most likely living fossils according to EPI than was the case for pEPI.
Improvements to Change
There was a significant correlation between the change variable and pEPI. The strength of the correlation was due to the correlation between the change and success variable. Under a punctuated equilibrium model of evolution in which most character evolution occurs during speciation events (Gould and Eldredge, 1993), we should expect there to be a correlation between taxic diversity and observed morphological/ecological changes. Although there is evidence that speciation events can lead to morphological changes (Venditti and Pagel, 2010; Ezard et al., 2013; Rabosky et al., 2013), the correlation between change and success can be alternatively explained by there being more opportunity to observe changes in more speciose clades: character changes may remain hidden along long solitary branches as there are no taxa to indicate any possibility of change. In our calculation of the change metric we attempted to control for this by weighting the amount of observed change by the total branch length represented by the clade. An alternative approach may instead be to count the number of uniquely derived characters (autapomorphies) that have arisen for a given clade, as these characters are by definition independent of the number of species in a clade (Cavin and Guinot, 2014). For such a large-scale study as ours, however, an automated method of identifying autapomorphic characters would need to be developed. Furthermore, it is unlikely that switching to an autapomorphic character change variable would impact the overall rankings because, as indicated in the results, the EPI character was mostly impacted by the time variable.
Progressivism and the Living Fossil
Amemiya et al. (2013) recently demonstrated that the coelacanth lineage has experienced reduced rates of evolution in protein-coding sequences relative to other vertebrate lineages; implicitly reinforcing the status of coelacanths as living fossils. Equating little change at the molecular scale as evidence for the living fossil status for coelacanths, however, has led to many rebuttals (Bockmann et al., 2013; Chalopin et al., 2014; Forconi et al., 2014; Naville et al., 2015). In particular, its publication and the publication of similar articles have reignited arguments over the need to avoid ‘progressivist’ language in evolutionary discourse (Casane and Laurenti, 2013; Grandcolas et al., 2014; Minelli and Baedke, 2014). As discussed above, under a progressivist or gradist interpretation of evolution the term living fossil may be interpreted to mean a ‘low-ranked’ or ‘basal’ taxon that is more closely related to an extinct ancestor than any ‘high-ranked’ taxa. It is argued that the usage of such progressivist language as living fossil is detrimental as it can lead to the promotion of this false interpretation of evolution, particularly among the public (Rigato and Minelli, 2013). It should be highlighted, however, that our use of the term living fossil is based on a phylogenetic approach: we only consider monophyletic clades and use sister-comparisons. In no way is our use of the term ‘progressivist’. We developed metrics that measured evolutionary activity relative to a clade’s sister, and our interpretation of an ‘evolutionarily low performing clade’ is not distinct from similar, recent studies that have re-appraised the status of well-known living fossils (Cavin and Guinot, 2014; Herrera-Flores et al., 2017). Do we consider taxa that have scored low on our EPI/pEPI metrics as true living fossils? Ultimately, this is an essentialist question, but we consider the use of the term in conjunction with our metrics, as a synonym for a clade experiencing little evolutionary output, to be of potential utility.
CONCLUSIONS
The performance of ED, pEPI and EPI metrics (measured against our Wikipedia-sourced measure of popular usage) is similar, but our metrics (EPI and pEPI) have many practical advantages. By covering all six identified aspects of the living fossil, pEPI and EPI capture more elements of the living fossil concept. Additionally, it is possible to calculate our metrics above the species-level using data that can be relatively easily sourced. pEPI does not require a fully resolved time-calibrated phylogenetic tree because estimates of success can be determined from online taxonomies, and large available datasets of “time since split” are also now available online. EPI is more demanding of information in that it requires a sufficient number of character states and a phylogenetic tree in order to infer ancestral states. This tree, however, need not be time-calibrated nor fully resolved. Additionally, sister contrasts result in there being less need to ensure that characters selected are representative of a group as a whole; instead, they need only be comparable with the sister.
In summary, EPI and pEPI can be readily calculated from taxonomy, time since divergence and, optionally, character matrices; they provide the most reliable means available of quantifying the degree to which a clade fits the “living fossil” concept. These metrics enable the living fossil concept to be investigated in a more quantitative manner, and hence to provide a better grounding for investigations of the reality, significance, and potential causes of the phenomenon.
ACKNOWLEDGEMENTS
The authors wish to thank A. Purvis and L.H. Liow for early suggestions of ideas; F.R. Leiva, J. Bryant, S. Dures and J. Hansford for useful discussion. In addition, we wish to thank L. Cavin and three anonymous reviewers whose insightful comments and suggestions have greatly improved this article.
REFERENCES
Alfaro, M.E., Santini, F., Brock, C., Alamillo, H., Dornburg, A., Rabosky, D.L., Carnevale, G., and Harmon, L.J. 2009. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proceedings of the National Academy of Sciences of the United States of America, 106(32):13410-13414. https://doi.org/10.1073/pnas.0811087106
Amemiya, C.T., Alföldi, J., Lee, A.P., Fan, S., Philippe, H., Maccallum, I., Braasch, I., Manousaki, T., Schneider, I., Rohner, N., Organ, C., Chalopin, D., Smith, J.J., Robinson, M., Dorrington, R.A., Gerdol, M., Aken, B., Biscotti, M.A., Barucca, M., Baurain, D., Berlin, A.M., Blatch, G.L., Buonocore, F., Burmester, T., Campbell, M.S., Canapa, A., Cannon, J.P., Christoffels, A., De Moro, G., Edkins, A.L., Fan, L., Fausto, A.M., Feiner, N., Forconi, M., Gamieldien, J., Gnerre, S., Gnirke, A., Goldstone, J.V., Haerty, W., Hahn, M.E., Hesse, U., Hoffmann, S., Johnson, J., Karchner, S.I., Kuraku, S., Lara, M., Levin, J.Z., Litman, G.W., Mauceli, E., Miyake, T., Mueller, M.G., Nelson, D.R., Nitsche, A., Olmo, E., Ota, T., Pallavicini, A., Panji, S., Picone, B., Ponting, C.P., Prohaska, S.J., Przybylski, D., Saha, N.R., Ravi, V., Ribeiro, F.J., Sauka-Spengler, T., Scapigliati, G., Searle, S.M.J., Sharpe, T., Simakov, O., Stadler, P.F., Stegeman, J.J., Sumiyama, K., Tabbaa, D., Tafer, H., Turner-Maier, J., van Heusden, P., White, S., Williams, L., Yandell, M., Brinkmann, H., Volff, J.-N., Tabin, C.J., Shubin, N., Schartl, M., Jaffe, D.B., Postlethwait, J.H., Venkatesh, B., Di Palma, F., Lander, E.S., Meyer, A. and Lindblad-Toh, K. 2013. The African coelacanth genome provides insights into tetrapod evolution. Nature, 496(7445):311-6. https://doi.org/10.1038/nature12027
AmphibiaWeb. 2016. Amphibia Web, read number of species November 2016. University of California, Berkeley, CA, USA. http://www.amphibiaweb.org
Baillie, J.E.M., Hilton-Taylor, C., and Stuart, S.N. 2004. 2004 IUCN red list of threatened species: a global species assessment. International Union for Conservation of Nature.
Bennett, D.J. 2016. Project-EPI: quantifying the living fossil with an evolutionary performance index (EPI). https://github.com/DomBennett/Project-EPI
Bennett, D.J., Sutton, M.D., and Turvey, S.T. 2017. Treeman: An R package for efficient and intuitive manipulation of phylogenetic trees. BMC Research Notes, 10(1):30. https://doi.org/10.1186/s13104-016-2340-8
Bininda-Emonds, O.R.P., Cardillo, M., Jones, K.E., MacPhee, R.D.E., Beck, R.M.D., Grenyer, R., Price, S.A., Vos, R.A., Gittleman, J.L., and Purvis, A. 2007. The delayed rise of present-day mammals. Nature, 446(7135):507-12. https://doi.org/10.1038/nature05634
Birdtree. 2016. A global phylogeny of birds, trees downloaded in 2014. birdtree.org
Biton, R., Geffen, E., Vences, M., Cohen, O., Bailon, S., Rabinovich, R., Malka, Y., Oron, T., Boistel, R., Brumfeld, V., and Gafny S. 2013. The rediscovered Hula painted frog is a living fossil. Nature Communications, 4:1959. https://doi.org/10.1038/ncomms2959
Bockmann, F.A., De Carvalho, M.R., and De Carvalho, M. 2013. The salmon, the lungfish (or the coelacanth) and the cow: a revival? Zootaxa, 3750(3):265. https://doi.org/10.11646/zootaxa.3750.3.6
BOLD. 2016. Barcode of life database: insecta {class}, statistics read from left-hand column in November 2016. http://www.boldsystems.org/index.php/TaxBrowser_Taxonpage?taxid=82
Buckley, G., Brochu, C., Krause, D.W., and Pol, D. 2000. A pug-nosed crocodyliform from the Late Cretaceous of Madagascar. Nature, 405(6789):941-4. https://doi.org/10.1038/35016061
Casane, D. and Laurenti, P. 2013. Why coelacanths are not “living fossils”: a review of molecular and morphological data. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 35(4):332-8. https://doi.org/10.1002/bies.201200145
Cavin, L. and Guinot, G. 2014. Coelacanths as “almost living fossils.” Frontiers in Ecology and Evolution, 2(49):1-5. https://doi.org/10.3389/fevo.2014.00049
Cavin, L. and Kemp, A. 2011.The impact of fossils on the evolutionary distinctiveness and conservation status of the Australian lungfish. Biological Conservation, 144(12):3140-3142. https://doi.org/10.1016/j.biocon.2011.08.014
Chalopin, D., Fan, S., Simakov, O., Meyer, A., Schartl, M., and Volff, J.N. 2014. Evolutionary active transposable elements in the genome of the coelacanth. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 322(6):322-333. https://doi.org/10.1002/jez.b.22521
Cieslak, A., Fresneda, J., and Ribera, I. 2014. Life-history specialization was not an evolutionary dead-end in Pyrenean cave beetles. Proceedings of the Royal Society of London B - Biological Sciences, 281(1781):20132978. https://doi.org/10.1098/rspb.2013.2978
Darwin, C. 1859. On the Origin of Species by Means of Natural Selection. John Murray, London.
Eldredge, N. 1979. Alternative approaches to evolutionary theory, p. 7-19. In Schwartz, J.H. and Rolins, H.B. (eds.), Models and methodologies in evolutionary theory. Bulletin of the Carnegie Museum of Natural History.
Eldredge, N. 1984. Simpson’s inverse: bradytely and the phenomenon of living fossils, p. 272-277. In Eldredge, N. and Stanley, S.M. (eds.), Living Fossils (Casebooks in Earth Sciences), Springer-Verlag, New York.
Eldredge, N. and Stanley, S.M. 1984. Living Fossils (Casebooks in Earth Sciences). Springer-Verlag, New York. https://doi.org/10.1007/978-1-4613-8271-3
Ezard, T.H.G., Thomas, G.H., and Purvis, A. 2013. Inclusion of a near-complete fossil record reveals speciation-related molecular evolution. Methods in Ecology and Evolution, 4(8):745-753. https://doi.org/10.1111/2041-210X.12089
Federhen, S. 2012. The NCBI taxonomy database. Nucleic Acids Research, 40(Database issue): D136-43. https://doi.org/10.1093/nar/gkr1178
Fisher, D.C. 1990. Rates of evolution - living fossils, p. 152-159. In Briggs, D.E.G. and Crowther, P.R. (eds.), Paleobiology: A Synthesis. Blackwell Science, Oxford.
Forconi, M., Chalopin, D., Barucca, M., Biscotti, M.A., De Moro, G., Galiana, D., Gerdol, M., Pallavicini, A., Canapa, A., Olmo, E., and Volff, J.N. 2014. Transcriptional activity of transposable elements in coelacanth. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 322(6):379-389. https://doi.org/10.1002/jez.b.22527
Froese, R. and Pauly, D. 2017. FishBase, looked up number of species November 2017. fishbase.org
Garcia-Fernàndez, J. and Benito-Gutiérrez, É. 2009. It’s a long way from amphioxus: descendants of the earliest chordate. BioEssays, 31(6):665-675. https://doi.org/10.1002/bies.200800110
Gill, F.B. and Wright, M.T. 2006. Birds of the World: Recommended English Names, Princeton University Press, Princeton.
Gould, S.J. and Eldredge, N. 1993. Punctuated equilibrium comes of age. Nature, 366(6452):223-227. https://doi.org/10.1038/366223a0
Grandcolas, P., Nattier, R., and Trewick, S. 2014. Relict species: a relict concept? Trends in Ecology and Evolution, 29(12):655-663. https://doi.org/10.1016/j.tree.2014.10.002
Hackett, S.J., Kimball, R.T., Reddy, S., Bowie, R.C.K., Braun, E.L., Braun, M.J., Jena L., Chojnowski, J.L., Cox, W.A, Han, K.-L., Harshman, J., Huddleston, C.J., Marks, B.D., Miglia, K.J., Moore, W.S., Sheldon, F.H., Steadman, D.W. Witt, C.C., and Yuri, T. 2008. A phylogenomic study of birds reveals their evolutionary history. Science, 320(5884):1763-8. https://doi.org/10.1126/science.1157704
Hanazawa, M., Narushima, H., and Minaka, N. 1995. Generating most parsimonious reconstructions on a tree: a generalization of the Farris-Swofford-Maddison method. Discrete Applied Mathematics, 56:245-265. https://doi.org/10.1016/S0166-218X(97)00088-7
Hastie, T.J. and Pregibon, D. 1992. Generalized linear models, chapter 6. In Chambers, J.M. and Hastie, T.J. (eds.), Statistical Models in S., Wadsworth & Brooks/Cole Advanced Books & Software.
Hay, J.M., Subramanian, S., Millar, C.D., Mohandesan, E., and Lambert, D.M. 2008. Rapid molecular evolution in a living fossil. Trends in Genetics, 24:106-109. https://doi.org/10.1016/j.tig.2007.12.002
Hedges S.B., Dudley J., and Kumar S. 2006. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics, 22:2971-2972. https://doi.org/10.1093/bioinformatics/btl505
Hedges S.B., Marin J., Suleski M., Paymer, M., and Kumar S. 2015. Tree of Life Reveals Clock-Like Speciation and Diversification. Molecular Biology and Evolution, 32:835-845. https://doi.org/10.1093/molbev/msv037
Herrera-Flores, J.A., Stubbs, T.L., and Benton, M.J. 2017. Macroevolutionary patterns in Rhynchocephalia: is the tuatara (Sphenodon punctatus) a living fossil? Palaeontology, 1-10. https://doi.org/10.1111/pala.12284
Isaac, N.J.B., Turvey, S.T., Collen, B., Waterman, C., and Baillie, J.E.M. 2007. Mammals on the EDGE: conservation priorities based on threat and phylogeny. PloS One, 2(3):e296. https://doi.org/10.1371/journal.pone.0000296
ITIS. 2016. Integrated Taxonomic Information System Database. https://www.itis.gov
Jackson, J.B.C. 1974. Biogeographic consequences of eurytopy and stenotopy among marine bivalves and their evolutionary significance. The American Naturalist, 108(962):541-560. https://doi.org/10.1086/282933
Jetz, W., Thomas, G.H.H., Joy, J.B.B., Hartmann, K., and Mooers, A.O. 2012. The global diversity of birds in space and time. Nature, 491(7424):1-5. https://doi.org/10.1038/nature11631
Jones, K.E., Bielby, J., Cardillo, M., Fritz, S.A., O’Dell, J., Orme, C.D.L., Safi, K., Sechrest, W., Boakes, E.H., Carbone, C., Connolly, C., Cutts, M.J., Foster, J.K., Grenyer, R., Habib, M., Plaster, C.A., Price, S.A. Rigby, E.A., Rist, J., Teacher, A., Bininda-Emonds, O.R.P. Gittleman, J.L., Mace, G.M., and Purvis, A. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology, 90:2648-2648. https://doi.org/10.1890/08-1494.1
Kano, Y. Kimura, S., Kimura, T., and Warén, A. 2012. Living Monoplacophora: morphological conservatism or recent diversification? Zoologica Scripta, 41(5):471-488. https://doi.org/10.1111/j.1463-6409.2012.00550.x
Kin, A. and Błazejowski, B. 2014. The horseshoe crab of the genus Limulus : living fossil or stabilomorph? PLoS One, 9(10). https://doi.org/10.1371/journal.pone.0108036
Kristensen, R.M. 2002. An introduction to loricifera, cycliophora, and micrognathozoa. Integrative and Comparative Biology, 42(3):641-51. https://doi.org/10.1093/icb/42.3.641
Kumar, S. and Hedges, S.B. 2011. TimeTree2: species divergence times on the iPhone. Bioinformatics, 27:2023-2024. https://doi.org/10.1093/bioinformatics/btr315
Lislevand, T., Figuerola, J., and Székely, T. 2007. Avian body sizes in relation to fecundity, mating system, display behavior, and resource sharing. Ecology, 88(12):3218. https://doi.org/10.1890/06-2054
Livezey, B.C. and Zusi, R.L. 2007. Higher-order phylogeny of modern birds (Theropoda, Aves: Neornithes) based on comparative anatomy. II. Analysis and discussion. Zoological Journal of the Linnean Society, 149(1):1-95. https://doi.org/10.1111/j.1096-3642.2006.00293.x
Mathers, T.C., Hammond, R.L., Jenner, R.A., Hänfling, B., and Gómez, A. 2013. Multiple global radiations in tadpole shrimps challenge the concept of “living fossils”. PeerJ, 1:e62. https://doi.org/10.7717/peerj.62
Meloro, C. and Jones, M.E.H. 2012. Tooth and cranial disparity in the fossil relatives of Sphenodon (Rhynchocephalia) dispute the persistent “living fossil” label. Journal of Evolutionary Biology, 25(11):2194-2209. https://doi.org/10.1111/j.1420-9101.2012.02595.x
Minelli, A. and Baedke, J. 2014. Model organisms in evo-devo: promises and pitfalls of the comparative approach. History and Philosophy of the Life Sciences, 36(1):42-59. https://doi.org/10.1007/s40656-014-0004-3
Nagalingum, N.S., Marshall, C.R., Quental, T.B., Rai, H.S., Little, D.P., and Mathews, S. 2011. Recent synchronous radiation of a living fossil. Science, 334(6057):796-799. https://doi.org/10.1126/science.1209926
Naville, M., Chalopin, D., Casane, D., Laurenti, P., and Volff, J.N. 2015. The coelacanth: can a “living fossil” have active transposable elements in its genome? Mobile Genetic Elements, 5(4):55-59. https://doi.org/10.1080/2159256X.2015.1052184
Near, T.J., Dornburg, A., Tokita, M., Suzuki, D., Brandley, M.C., and Friedman, M. 2014. Boom and bust: ancient and recent diversification in bichirs (Polypteridae: Actinopterygii), a relictual lineage of ray-finned fishes. Evolution, 68(4):1014-1026. https://doi.org/10.1111/evo.12323
O’Leary, M.A., Bloch, J.I., Flynn, J.J., Gaudin, T.J., Giallombardo, A., Giannini, N.P., Goldberg, S.L., Kraatz, B.P., Luo, Z.-X., Meng, J., Ni, X., Novacek, M.J., Perini, F.A., Randall, Z.S., Rougier, G.W., Sargis, E.J., Silcox, M.T., Simmons, N.B., Spaulding, M., Velazco, P.M., Weksler, M., Wible, J.R., and Cirranello, A.L. 2013. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science, 339(6120):662-7. https://doi.org/10.1126/science.1229237
O’Reilly, J.E. Puttick, M.N., Parry, L., Tanner, A.R., Tarver, J.E., Fleming, J., Pisani, D., and Donoghue, P.C.J. 2016. Bayesian methods outperform parsimony but at the expense of precision in the estimation of phylogeny from discrete morphological data. Biology Letters, 12(4):1-5. https://doi.org/10.1098/rsbl.2016.0081
Obst, M., Faurby, S. Bussarawit, S., and Funch, P. 2012. Molecular phylogeny of extant horseshoe crabs (Xiphosura, Limulidae) indicates Paleogene diversification of Asian species. Molecular Phylogenetics and Evolution, 62(1):21-6. https://doi.org/10.1016/j.ympev.2011.08.025
Ødegaard, F. 2000. How Many Species of Arthropods? Erwin’s Estimate Revised. Biological Journal of the Linnean Society, 71(4):583-597. https://doi.org/10.1111/j.1095-8312.2000.tb01279.x
Paradis, E., Claude, J., and Strimmer, K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics, 20(2):289-290. https://doi.org/10.1093/bioinformatics/btg412
Rabosky, D.L., Santini, F., Eastman, J., Smith, S.A., Sidlauskas, B., Chang, J., and Alfaro, M.E. 2013. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nature Communications, 4(1958):1-8. https://doi.org/10.1038/ncomms2958
Rigato, E. and Minelli, A. 2013. The great chain of being is still here. Evolution: Education and Outreach, 6(1):8. https://doi.org/10.1186/1936-6434-6-18
Royer, D.L., Hickey, L.J., and Wing, S.L. 2003. Ecological conservatism in the “living fossil” ginkgo. Paleobiology, 29(1):84-104. https://doi.org/10.1666/0094-8373(2003)029<0084:ECITLF>2.0.CO;2
Royer-Carenzi, M., Pontarotti, P., and Didier, G. 2013. Choosing the best ancestral character state reconstruction method. Mathematical Biosciences, 242(1):95-109. https://doi.org/10.1016/j.mbs.2012.12.003
Rudkin, D.M., Young, G.A., and Nowlan, G.S. 2008. The oldest horseshoe crab: a new xiphosurid from late Ordovician Konservat-Lagerstätten deposits, Manitoba, Canada. Palaeontology, 51(1):1-9. https://doi.org/10.1111/j.1475-4983.2007.00746.x
Schierwater, B., Eitel, M., Jakob, W., Osigus, H.J., Hadrys, H., Dellaporta, S.L., Kolokotronis, S.O., and DeSalle, R. 2009. Concatenated analysis sheds light on early metazoan evolution and fuels a modern “urmetazoon” hypothesis. PLoS Biology, 7(1). https://doi.org/10.1371/journal.pbio.1000020
Schopf, T.J.M. 1984. Rates of evolution and the notion of “living fossils.” Annual Review of Earth Planetary Science, 12:245-292. https://doi.org/10.1146/annurev.ea.12.050184.001333
Smith, J.L.B. 1939. A living fish of Mesozoic type. Nature, 143(3620):455-456. https://doi.org/10.1038/143455a0
Smith, J.J., Sumiyama, K., and Amemiya, C.T. 2012. A living fossil in the genome of a living fossil: Harbinger transposons in the coelacanth genome. Molecular Biology and Evolution, 29(3):985-993. https://doi.org/10.1093/molbev/msr267
Stanley, S.M. 1998. Macroevolution: Pattern and Process. The Johns Hopkins University Press, Baltimore.
Suzuki, T.G., Ogino, K., Tsuneki, K., and Furuya, H. 2010. Phylogenetic analysis of dicyemid mesozoans (phylum Dicyemida) from innexin amino acid sequences: dicyemids are not related to Platyhelminthes. The Journal of Parasitology, 96(3):614-625. https://doi.org/10.1645/GE-2305.1
Syed T. and Schierwater, B. 2002. The evolution of the Placozoa: a new morphological model. Senckenbergiana Lethaea, 82(1):315-324. https://doi.org/10.1007/BF03043791
The Plant List. 2013. A Working List of Plants (Version 1.1.), looked up number of accepted names November 2016. http://www.theplantlist.org/
Thomas, G.H., Cooper, N., Venditti, C., Meade, A., and Freckleton, R.P. 2014. Bias and measurement error in comparative analyses: a case study with the Ornstein Uhlenbeck model. bioRxiv. https://doi.org/10.1101/004036
Uetz, P. 2010. The Original Descriptions of Reptiles. Zootaxa, 68(2334):59-68.
Venditti, C. and Pagel, M. 2010. Speciation as an active force in promoting genetic evolution. Trends in Ecology and Evolution, 25(1):14-20. https://doi.org/10.1016/j.tree.2009.06.010
Venkatesh, B., Lee, A.P., Ravi, V., Maurya, A.K., Lian, M.M., Swann, J.B., Ohta, Y., Flajnik, M.F., Sutoh, Y., Kasahara, M., Hoon, S., Gangu, V., Roy, S.W., Irimia, M., Korzh, V., Kondrychyn, I., Lim, Z.W., Tay, B.-H., Tohari, S., Kong, K.W., Ho, S., Lorente-Galdos, B., Quilez, J., Marques-Bonet, T., Raney, B.J., Ingham, P.W., Tay, A., Hillier, L.W., Minx, P., Boehm, T., Wilson, R.K., Brenner, S., Warren, W.C. 2014. Elephant shark genome provides unique insights into gnathostome evolution. Nature, 505(7482):174-9. https://doi.org/10.1038/nature12826
Voigt, O., Collins, A.G., Pearse, V.B., Pearse, J.S., Ender, A., Hadrys, H., and Schierwater, B. 2004. Placozoa - no longer a phylum of one. Current Biology, 14(22):R944-R945. https://doi.org/10.1016/j.cub.2004.10.036
Vrba, E. 1984. Evolutionary pattern and process in the sister-group Alcelaphini-Aepycerotini (Mammalia: Bovidae), p. 62-79. In Eldredge, N. and Stanley, S.M. (eds.), Living Fossils (casebooks in Earth Sciences), Springer-Verlag, New York.
Webster, A. and Purvis, A. 2002. Testing the accuracy of methods for reconstructing ancestral states of continuous characters. Proceedings of the Royal Society B: Biological Sciences, 269(1487):143-149. https://doi.org/10.1098/rspb.2001.1873
Wikipedia. 2016. Wikipedia: The Free Enclycopedia, en.wikipedia.org
Wilson, D.E. and Reeder, D.M. 2005. Mammal Species of the World. Johns Hopkins University Press, Baltimore.
Yoshida, K. 2002. Long survival of “living fossils” with low taxonomic diversities in an evolving food web. Paleobiology, 28(4):464-473. https://doi.org/10.1666/0094-8373(2002)028<0464:LSOLFW>2.0.CO;2

