 Mammoths, molecules and morphology: A case study in ancient speciation
Mammoths, molecules and morphology: A case study in ancient speciation
Article number: 27.3.a52
https://doi.org/10.26879/1419
Copyright Palaeontological Association, October 2024
Author biographies
Plain-language and multi-lingual abstracts
PDF version
Submission: 7 June 2024. Acceptance: 14 October 2024.
ABSTRACT
Recent research on ancient DNA back to a million years or more has contributed significantly to our understanding of mammalian evolution. We focus on mammoths, the most intensively studied lineage after humans. Recent genomic data confirm the model, originally based on morphology, that Early Pleistocene mammoths from northeast Siberia, of relatively advanced morphology, were the source of endemic North American mammoth species and the Holarctic woolly mammoth. The new data further reveal at least two introgression events in the origin of the Late Pleistocene North American mammoth, resulting from hybridisation between the endemic North American form and later immigrating woolly mammoth populations. This exemplar highlights broader implications for the reconciliation of molecular and morphological data in palaeontology. These include the detection of non-bifurcating phylogenetic processes such as hybrid speciation; the problem of morphologically cryptic species identified from DNA; and the issues these raise for species concepts, taxonomy and nomenclature of fossil taxa.
Adrian M. Lister. Natural History Museum, Cromwell Road, London SW7 5BD, UK. Corresponding author. a.lister@nhm.ac.uk
Love Dalén. Centre for Palaeogenetics, Svante Arrhenius väg 20C, SE-106 91 Stockholm, Sweden; Department of Zoology, Stockholm University, Sweden. love.dalen@zoologi.su.se
Keywords: mammoths; Mammuthus columbi; molar morphology; ancient DNA; hybrid speciation; species concepts
Final citation: Lister, Adrian M. and Dalén, Love. 2024. Mammoths, molecules and morphology: A case study in ancient speciation. Palaeontologia Electronica, 27(3):a52.
https://doi.org/10.26879/1419
palaeo-electronica.org/content/2024/5360-mammoths-dna-and-morphology
Copyright: October 2024 Palaeontological Association.
This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
creativecommons.org/licenses/by/4.0
INTRODUCTION
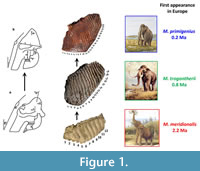 An early success of ancient DNA research was the resolution of the phylogenetic position of the woolly mammoth Mammuthus primigenius: it was more closely related to the living Asian elephant Elephas maximus than to the African elephants Loxodonta spp. (Rohland et al., 2010). The fossil record demonstrates that this diversification had occurred within Africa by about 5 Ma (million years ago), and that by 3.5-3.0 Ma mammoths had dispersed to Europe and the Far East (Lister et al., 2005; Wei et al., 2006). The European record shows a series of species through the Plio-Pleistocene, from Mammuthus rumanus to M. meridionalis (‘southern mammoth’), to M. trogontherii (‘steppe mammoth’) to M. primigenius (woolly mammoth), with substantial changes including shortening and deepening of the skull and increase in the crown height and number of enamel ridges in the molars (Figure 1). Mammoths dispersed to North America some time 1.5-1.2 Ma, with various taxonomic names having been applied to early forms but Mammuthus columbi (Columbian mammoth) accepted as the common species of the Late Pleistocene (Lister, 2017).
An early success of ancient DNA research was the resolution of the phylogenetic position of the woolly mammoth Mammuthus primigenius: it was more closely related to the living Asian elephant Elephas maximus than to the African elephants Loxodonta spp. (Rohland et al., 2010). The fossil record demonstrates that this diversification had occurred within Africa by about 5 Ma (million years ago), and that by 3.5-3.0 Ma mammoths had dispersed to Europe and the Far East (Lister et al., 2005; Wei et al., 2006). The European record shows a series of species through the Plio-Pleistocene, from Mammuthus rumanus to M. meridionalis (‘southern mammoth’), to M. trogontherii (‘steppe mammoth’) to M. primigenius (woolly mammoth), with substantial changes including shortening and deepening of the skull and increase in the crown height and number of enamel ridges in the molars (Figure 1). Mammoths dispersed to North America some time 1.5-1.2 Ma, with various taxonomic names having been applied to early forms but Mammuthus columbi (Columbian mammoth) accepted as the common species of the Late Pleistocene (Lister, 2017).
The evolution of Eurasian mammoths was generally considered as the transformation of a single lineage, for example as envisaged by Maglio (1973) (Figure 1). This picture was radically altered by the discovery of trogontherii -like mammoths in the Early Olyorian (ca. 1.2 Ma) of NE Siberia, and even earlier (ca. 1.7 Ma) in NE China, much earlier than the first appearance of Mammuthus trogontherii in Europe. These mammoths presumably evolved from eastern populations of M. meridionalis which it had replaced in eastern Asia by these dates (Lister and Sher, 2001, 2015; Wei et al., 2010). These studies thus suggested that European Mammuthus trogontherii arrived by westward dispersal from eastern Asia, displacing and/or introgressing with endemic M. meridionalis.
For the North American mammoth Mammuthus columbi, several models had been proposed, all of them assuming dispersal from Asia across Beringia:
- that only Mammuthus meridionalis dispersed there, evolving (perhaps through a ‘ Mammuthus imperator ’ intermediate) into M. columbi (Maglio, 1973);
- that both meridionalis and trogontherii mammoths dispersed there, the former being a ‘dead end’, the latter being the origin of M. columbi (Osborn, 1942; Harington, 1984; McDaniel and Jefferson, 2003);
- that only M. trogontherii dispersed there, being very similar to, and the immediate ancestor of, M. columbi (Lister and Sher 2001, 2015; see Figure 2).
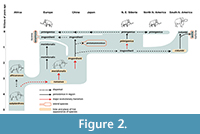 Support for the third hypothesis was provided by Lister and Sher (2015) based on (i) the replacement of M. trogontherii in eastern Asia by at least 1.7 Ma, before the dispersal into North America, (ii) the very similar morphology of M. trogontherii and M. columbi, and (iii) the absence of fossils of M. meridionalis grade in Beringia or North America.
Support for the third hypothesis was provided by Lister and Sher (2015) based on (i) the replacement of M. trogontherii in eastern Asia by at least 1.7 Ma, before the dispersal into North America, (ii) the very similar morphology of M. trogontherii and M. columbi, and (iii) the absence of fossils of M. meridionalis grade in Beringia or North America.
The earliest woolly mammoth Mammuthus primigenius was identified in the Late Olyorian (ca. 0.8-0.6 Ma) of NE Siberia and was proposed as the origin of European and North American M. primigenius by westward and eastward dispersal, respectively, in the Middle to Late Pleistocene (Lister and Sher, 2001, 2015). The pattern of transformation of mammoth molars in Europe through the late Middle Pleistocene suggests gene flow or introgression from immigrating M. primigenius into endemic M. trogontherii, rather than simple replacement (Lister, 2022).
Recent DNA analysis by van der Valk et al. (2021) has corroborated several aspects of this model, including the origin of endemic North American mammoths and, later, the woolly mammoth, from trogontherii-like Siberian ancestors. However, the genomic data also revealed previously unsuspected complexity, including a deep genetic division within the Early Pleistocene mammoths of NE Siberia, and a major hybridisation event in Middle Pleistocene mammoths of North America. These findings raise important issues of taxonomy, and evolution, here to be discussed.
GENOMIC INSIGHTS INTO MAMMOTH EVOLUTION
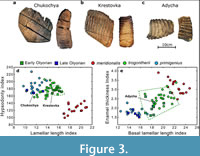 Abundant mammoth fossils were excavated by Sher (1986) from the Olyorian deposits of the Yana-Kolyma lowland of NE Siberia. Based on faunal and palaeomagnetic evidence he divided the Olyorian into two stratigraphical units, Early and Late Olyorian (stratigraphically Lower and Upper Olyorian), and initially described the mammoths as, respectively, Arctelephas sp. 1 and Arctelephas sp. 2 (Sher, 1986). These were subsequently subsumed within Mammuthus, the early ones (ca. 1.2-0.8 Ma) recognised as similar to European M. trogontherii while the late ones (ca. 0.8-0.5 Ma) approached the form of Late Pleistocene woolly mammoths, M. primigenius (Lister and Sher, 2001, 2015; Figure 3). The ancient DNA study (van der Valk et al., 2021) deliberately selected molars for analysis that morphologically fell clearly into one or the other group of Olyorian material. The Krestovka and Adycha specimens are Early Olyorian and trogontherii-like; the Chukochya specimen is Late Olyorian and primigenius-like (Figure 3). Figure 4 is a simplified representation of the major findings of the study, which will be dealt with in sequence.
Abundant mammoth fossils were excavated by Sher (1986) from the Olyorian deposits of the Yana-Kolyma lowland of NE Siberia. Based on faunal and palaeomagnetic evidence he divided the Olyorian into two stratigraphical units, Early and Late Olyorian (stratigraphically Lower and Upper Olyorian), and initially described the mammoths as, respectively, Arctelephas sp. 1 and Arctelephas sp. 2 (Sher, 1986). These were subsequently subsumed within Mammuthus, the early ones (ca. 1.2-0.8 Ma) recognised as similar to European M. trogontherii while the late ones (ca. 0.8-0.5 Ma) approached the form of Late Pleistocene woolly mammoths, M. primigenius (Lister and Sher, 2001, 2015; Figure 3). The ancient DNA study (van der Valk et al., 2021) deliberately selected molars for analysis that morphologically fell clearly into one or the other group of Olyorian material. The Krestovka and Adycha specimens are Early Olyorian and trogontherii-like; the Chukochya specimen is Late Olyorian and primigenius-like (Figure 3). Figure 4 is a simplified representation of the major findings of the study, which will be dealt with in sequence.
The Origin of the Woolly Mammoth Mammuthus primigenius
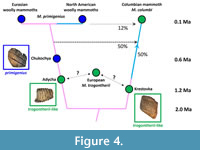 A major discovery of the DNA study is that Early Olyorian Adycha lineage of trogontherii-like morphology, the Late Olyorian ‘early primigenius ’ of Chukochya, and all later primigenius including European ones, are part of the same lineage (Figure 4). This finding is consistent with the hypothesis (Lister and Sher, 2001) that trogontherii -like Olyorian mammoths of NE Siberia included the progenitors of M. primigenius that then spread across the Holarctic. The evolutionary mode was not necessarily anagenetic, but could have entailed a series of cladogenetic events, since Adycha would in either case be equally genetically related to Chukochya and all Late Pleistocene primigenius.
A major discovery of the DNA study is that Early Olyorian Adycha lineage of trogontherii-like morphology, the Late Olyorian ‘early primigenius ’ of Chukochya, and all later primigenius including European ones, are part of the same lineage (Figure 4). This finding is consistent with the hypothesis (Lister and Sher, 2001) that trogontherii -like Olyorian mammoths of NE Siberia included the progenitors of M. primigenius that then spread across the Holarctic. The evolutionary mode was not necessarily anagenetic, but could have entailed a series of cladogenetic events, since Adycha would in either case be equally genetically related to Chukochya and all Late Pleistocene primigenius.
Two Clades of Early trogontherii-like Mammoths
Lister and Sher (2001, 2015) treated Early Olyorian trogontherii-like mammoths, dating to the interval 1.2-0.8 Ma, as a homogeneous entity, and the likely ancestor of European M. trogontherii. The study by Van der Valk et al. (2021) showed from nuclear DNA (nDNA) and mitochondrial DNA (mtDNA) that the two specimens from this assemblage, Adycha and Krestovka, are separate genetic lineages, despite being morphologically indistinguishable as far as we can tell from teeth. Median estimates for the date of their divergence from each other are 2.2 Ma and 2.3 Ma for mtDNA and nDNA, respectively.
Van der Valk et al. (2021) cautiously did not allocate taxonomic names to the Krestovka or Adycha lineages and only tentatively linked them to M. trogontherii. The species M. trogontherii was defined on European material of early Middle Pleistocene age (ca. 0.8-0.5 Ma) and there is currently no DNA data for it. Hence, we conservatively employ the descriptive term ‘trogontherii- like’ for the Siberian Early Olyorian forms to emphasise the morphological similarity without making any assumption about genetic relatedness. This term is equivalent to ‘trogontherioid’ in the study of Lister et al. (2005).
Nonetheless, consideration of the likely history of the Krestovka and Adycha lineages led van der Valk et al. (2021) to tentatively suggest the latter as the more likely ancestor, or close relative, of European M. trogontherii. The reason for this suggestion was evidence that the Krestovka lineage, after its occurrence in the Early Olyorian ca. 1.2-1.1 Ma, became extinct in Eurasia, leaving only the Adycha lineage. The D-statistics indicate an absence of gene flow between the Krestovka lineage and any of the Eurasian woolly mammoths (including Chukochya). Only the descendants of the Adycha lineage persist in the Late Olyorian and later deposits in Siberia, from there they would have dispersed westwards into Europe. Moreover, if the hypothesis by Lister and Sher (2015) that European Late Pleistocene woolly mammoths carried part of their ancestry from earlier European M. trogontherii is correct, then we could exclude the Krestovka lineage as ancestral to the European M. trogontherii, since this would have been manifested as excess allele sharing between the woolly mammoth from Scotland and Krestovka (van der Valk et al., 2021).
Considering the estimated divergence time between the Adycha and Krestovka lineages, the split between them took place roughly 1 Ma before the Krestovka individual lived (van der Valk et al., 2021). Hence there was a long period of contemporaneity between them, but their deep nuclear genomic divergence suggests that they were geographically separated. Both forms are allocated to the same geological unit in the same region (Lower Olyorian of the Yana-Kolyma lowland). However, as discussed in van der Valk et al. (2021, SI), while Krestovka is an in situ find at the base of the Early Olyorian (with an age constrained to 1.2–1.1 Ma), Adycha was an ex situ specimen circumstantially assigned to the first half of the Early Olyorian (1.2– 1.0 Ma) but with a possibly somewhat younger age, to 0.8 Ma (van der Valk et al., 2021, extended data figures 1 and 2 and supplementary text)). The DNA evidence also supports a somewhat younger age for the Adycha than the Krestovka specimen. We therefore consider it plausible that the Krestovka lineage became extinct in NE Siberia before 1.1 Ma and was replaced there by the Adycha lineage that expanded into NE Siberia from a currently unknown location.
Based on morphology and chronology, Mammuthus meridionalis, the Early Pleistocene mammoth species across mid-latitudes of Eurasia, is considered to be the ancestor of the trogontherii-like mammoths (Figure 2), but the genetic evidence for two lineages of trogontherii-like mammoths in Siberia opens several possible scenarios. M. meridionalis could have given rise to the common ancestor of Adycha and Krestovka, their trogontherii-like morphology evolving before their split (maybe in northern China where the earliest trogontherii-like mammoths are known). Conceivably, however, the split between Adycha and Krestovka could have happened during the meridionalis phase - before 1.7-2.0 Ma - and if so, their advanced morphology would have been a result of convergent evolution. That, if it could be demonstrated, would strengthen the case of their separate species status.
The Origin of North American Mammoths
The first entry date of mammoths into North America is constrained by absolute dating and/or good litho/biostratigraphic data at a small number of sites: Anza-Borrego, California (1.1-0.9 Ma), Leisey Shell Pit, Florida (ca. 1.3 Ma), Holloman’s Quarry, Frederick, Oklahoma (ca. 1.3 Ma), as well as Matanza Arroyo (1.4-1.3 Ma) and Adobe Ranch (1.6-1.2 Ma), both in New Mexico (source references in Agenbroad et al., 2005; Lister and Sher, 2015). In sum, we cannot reliably place it before 1.3 Ma, although an earlier date is possible.
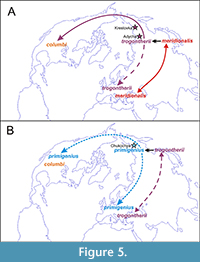 Various models for the evolution of North American mammoths have been proposed (see above), and these have led to differing taxonomies. Many authors assumed entry of a ‘primitive’, meridionalis-like form, either as precursor to M. columbi (Maglio, 1973) or a dead-end (Harington, 1984; McDaniel and Jefferson, 2003). According to this idea, early North American mammoths were either M. meridionalis itself, or a derivative North American endemic, retaining primitive features, such as ‘Mammuthus hayi’. Lister and Sher (2015) reviewed all such records and concluded that the fossils were either indeterminate or were morphologically indistinguishable from Late Pleistocene M. columbi, thereby proposing that all North American mammoths, from their first entry, derived from advanced, trogontherii -like mammoths of Eurasia (Figure 2 and Figure 5). This conclusion was supported by the similarity of North American mammoths, including some of the earliest ones, to those of the Early Olyorian, and the absence of any fossils attributable to M. meridionalis on either side of the Beringian transit route. All mainland North American mammoths could therefore be assigned to Mammuthus columbi until the Middle to Late Pleistocene entry of M. primigenius.
Various models for the evolution of North American mammoths have been proposed (see above), and these have led to differing taxonomies. Many authors assumed entry of a ‘primitive’, meridionalis-like form, either as precursor to M. columbi (Maglio, 1973) or a dead-end (Harington, 1984; McDaniel and Jefferson, 2003). According to this idea, early North American mammoths were either M. meridionalis itself, or a derivative North American endemic, retaining primitive features, such as ‘Mammuthus hayi’. Lister and Sher (2015) reviewed all such records and concluded that the fossils were either indeterminate or were morphologically indistinguishable from Late Pleistocene M. columbi, thereby proposing that all North American mammoths, from their first entry, derived from advanced, trogontherii -like mammoths of Eurasia (Figure 2 and Figure 5). This conclusion was supported by the similarity of North American mammoths, including some of the earliest ones, to those of the Early Olyorian, and the absence of any fossils attributable to M. meridionalis on either side of the Beringian transit route. All mainland North American mammoths could therefore be assigned to Mammuthus columbi until the Middle to Late Pleistocene entry of M. primigenius.
A key finding of recent genomic analysis is that Late Pleistocene North American M. columbi derive a substantial proportion of their DNA from the Krestovka lineage of NE Siberian trogontherii-like mammoths, indicating that this was likely the original lineage entering North America (van der Valk et al., 2021). This strongly supports the contention that the mammoths that seeded North America were already of advanced, ‘trogontherii’ type, as proposed on morphology by Lister and Sher (2015) (Figure 3). Our current DNA data is insufficient to estimate the split date between the Olyorian Krestovka specimen and Krestovka-like component in Late Pleistocene Columbian mammoths. If this were found to be in the region 1.5-1.0 Ma, it would lend additional support to early North American mammoths having derived from the Krestovka lineage.
In sum, genomic analysis confirms the suggestion, based on morphology and chronology, that the early trogontherii-like mammoths of NE Siberia were the precursors of both woolly (M. primigenius) and Columbian (M. columbi) mammoth species. However, unsuspected until the genomic analysis was the finding that these derived from already deeply divergent lineages, and that hybridisation played a significant role in the subsequent evolution of M. columbi, as will be discussed below.
The Role of Hybridisation
Migration and introgression. In the DNA study (van der Valk et al., 2021), a Late Pleistocene Mammuthus columbi individual showed an approximately 40/60 mix of Krestovka and ‘woolly mammoth’ DNA, respectively. A combination of admixture graph models and D-statistics suggested that this resulted from two or more waves of introgression with the endemic ‘Krestovka’ mammoths: the first, from a Middle Pleistocene ‘early primigenius’ lineage, brought in approximately 50%; the second, from Late Pleistocene M. primigenius, a further 11-13% (Figure 5). The Columbian mammoth’s mosaic genome is very unlikely to be the result of ancestral variation that pre-dated the Adycha/Krestovka split; this would not have produced the excess allele sharing observed between Krestovka and the Columbian mammoth and is incompatible with the bimodal distribution observed in ghost ancestry analysis (van der Valk et al., 2021).
Two separate phases of introgression were inferred because, for roughly 50% of its primigenius autosomal (nuclear) DNA, the Columbian mammoth is equally related to several clades of woolly mammoths (i.e., the hybridisation pre-dated much of primigenius diversification), whereas the extra ca. 12% shows a specific affinity to North American primigenius (i.e., the hybridisation post-dated primigenius diversification). The Mammuthus columbi DNA sample was from the ‘Union Pacific Mammoth’ from Chicken Springs, near Rawlins, Wyoming (UW6368), a male skeleton known as ‘Nip’ and with a radiocarbon age of ca. 13.4 cal ka, hence it post-dates both phases of hybridisation (Enk et al., 2016; Palkopoulou et al., 2018). Although van der Valk et al. (2021) tested only this one nuclear genome, we can have some confidence that it is representative of Late Pleistocene North American Columbian mammoths, i.e., they all had some level of hybrid ancestry. First, all tested Columbian mammoths are consistent in possessing a primigenius -derived mitogenome (Enk et al., 2016; Chang et al., 2017). Second, while the columbi individual with hybrid ancestry lived in the terminal Pleistocene, its hybrid autosomal genome arose much earlier. The hybrid genome could not have persisted in 50/50 or 60/40 proportion if it were in a small minority within a population of unhybridized individuals. Nonetheless, with limited geographic sampling, it is not impossible that some ‘pure Krestovka’ populations remained in North America even after the hybridisation event. There are large regions of mammoth range (such as the southern USA and Mexico) and long time periods for which we do not yet have any DNA data.
A Middle Pleistocene age of the first hybridisation is supported by the estimated most recent common ancestor of the introgressed mtDNA observed in all Columbian mammoths sequenced to date, with a median value of ca. 420 ka (95% range 511-338 ka). A hybrid origin based on mtDNA was earlier proposed by Enk et al. (2011, 2016), although they did not estimate the date of hybridisation but assumed it was Late Pleistocene as that is the earliest fossil evidence for entry of M. primigenius into North America. The 420 ka estimate should be considered a minimum age, since genetic drift (e.g., through a bottleneck) could have resulted in the time to most recent common ancestor (tmrca) being more recent than the actual hybridisation event. At the limit, it must be younger than 800 kya (the oldest possible age of the earliest known Siberian primigenius, represented by the Chukochya specimen), since the primigenius-like component of the Columbian mammoth’s genome falls inside the diversity of Chukochya (van der Valk et al., 2021, extended data fig. 7).
Studies of mitochondrial DNA (Enk et al., 2016; Chang et al., 2017) had identified two distinct lineages within North American Late Pleistocene mammoths. One of them includes all samples that had been identified morphologically as M. primigenius, and the other includes all samples that had been identified as M. columbi (plus two as ‘M. jeffersonii’: see below). As discussed above, the columbi lineage carries introgressed ‘primigenius’ mtDNA, and it is notable that within the global M. primigenius mtDNA tree the North American columbi and primigenius clades are sister groups (subclades of clade 1c), albeit divergent. This finding indicates that all of the mtDNA introgression occurred during the first hybridisation, in contrast to the nDNA where there was additional introgression during the second hybridisation. It also suggests the possibility that the two waves of primigenius dispersal into North America, and their hybridisation with endemic North American mammoths, came from the same, or closely related population(s) of woolly mammoth, separated by a substantial time interval during which mtDNA divergence had occurred.
We suggest two alternative models to explain this observation. Both begin with primigenius colonization of North America in the Middle Pleistocene, followed by hybridization with the resident ‘Krestovka’ mammoths to produce a hybridised columbi population that inhabited continental North America. In the first model, unhybridised primigenius persisted in Alaska/Yukon from ca 420 ka onwards, where it will have undergone genetic drift, and possibly some (male-mediated) gene flow from Siberia, making it genetically different from the 50% primigenius-like part of the columbi genome. In the Late Pleistocene, further hybridisation brought a second (11-13%) introgression from this population into native M. columbi in the south.
In the second model, the Middle Pleistocene primigenius population hybridized with the endemic ‘Krestovka’ population, but then went extinct. In the Late Pleistocene, a second wave of expansion from Siberia into North America occurred, leading to the second round of hybridization. This scenario is consistent with the nuclear genomic data (van der Valk et al., 2021, extended data fig. 7c). To explain the sister-group relationship between columbi and North American primigenius mtDNA within clade 1c, this model would predict that clade 1 resided during the early part of its history in easternmost Siberia, and both waves of expansion into North America came from a population carrying Clade 1c mtDNA.
It is, however, also possible that the proposed two hybridisation ‘events’ were in fact a sequence of intermittent hybridisations, perhaps as successive populations of primigenius (starting after the Late Olyorian) expanded across the Bering land connection at times of low sea level, or from a resident population in Eastern Beringia. These are interesting possibilites that could be tested with a larger genomic dataset.
Hybridisation and the fossil record. The models proposed above explain the origin of the north-south division between Late Pleistocene Mammuthus primigenius and M. columbi in North America. Some mammoth fossils, mostly from the US Midwest, have been named ‘Mammuthus jeffersonii’, and these have been suggested from their morphology to represent hybrid individuals (Enk et al., 2011; Lister, 2017). However, the entire columbi 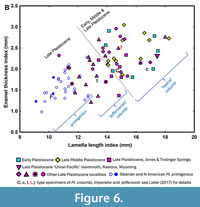 subclade (including ‘jeffersonii’ individuals) is now genomically identified as hybrid. For example, the genetically ‘double hybrid’ individual ‘Nip’ is entirely of columbi molar type (plate formula of last molar ×20p; lamellar frequency 5.5; enamel thickness 2.4 mm; Lister and Sher, 2015; Figure 6 herein), i.e., there is no trace of primigenius influence or even ‘jeffersonii’ morphology (thought to be intermediate between columbi and primigenius). It is, moreover, indistinguishable from Middle Pleistocene mammoth molars dated to ca. 400-200 ka (e.g., those from Mullen, Rushville & Hay Springs, Nebraska), that would have been subject to the first but not the second hybridisation event, and even from molars of Early Pleistocene mammoths such as those from Leisey Shell Pit, Florida, dated to ca. 1.3 Ma (Lister and Sher, 2015), that should have been subject to no hybridisation according to our model (Figure 6 and Figure 7). The first hybridisation event, therefore, imparted no visible morphological change, at least in the dentition of M. columbi and probably not in mandible form either (Lister and Sher, 2015, figs. S21, S24).
subclade (including ‘jeffersonii’ individuals) is now genomically identified as hybrid. For example, the genetically ‘double hybrid’ individual ‘Nip’ is entirely of columbi molar type (plate formula of last molar ×20p; lamellar frequency 5.5; enamel thickness 2.4 mm; Lister and Sher, 2015; Figure 6 herein), i.e., there is no trace of primigenius influence or even ‘jeffersonii’ morphology (thought to be intermediate between columbi and primigenius). It is, moreover, indistinguishable from Middle Pleistocene mammoth molars dated to ca. 400-200 ka (e.g., those from Mullen, Rushville & Hay Springs, Nebraska), that would have been subject to the first but not the second hybridisation event, and even from molars of Early Pleistocene mammoths such as those from Leisey Shell Pit, Florida, dated to ca. 1.3 Ma (Lister and Sher, 2015), that should have been subject to no hybridisation according to our model (Figure 6 and Figure 7). The first hybridisation event, therefore, imparted no visible morphological change, at least in the dentition of M. columbi and probably not in mandible form either (Lister and Sher, 2015, figs. S21, S24).
 Even the second proposed hybridisation, in the Late Pleistocene, produced ‘advanced’ mammoths of ‘jeffersonii’ type only sporadically (Lister, 2017). In the analysis by Chang et al. (Chang et al., 2017, fig. 1), of 16 North American mammoths of the Late Pleistocene, presumably therefore ‘double hybrids’, most had been morphologically classified as plain columbi (such as the ‘Nip’ individual), only two as ‘jeffersonii’. Moreover, Lister and Sher (2015, SI) found that samples of mammoth molars previously classified as ‘jeffersonii’ (such as those from Jones and Trolinger Springs, Missouri: Saunders, 1988) fall metrically within the range of Late Pleistocene mammoths referred to M. columbi (Lister, 2017, fig. 2; Figure 6 herein). Within both of these groups are individuals of more ‘primigenius- like’ morphology that could be informally termed ‘Jeffersonian’ if wished (Figure 6). Such individuals (including, for example, those described by Saunders et al., 2010) may perhaps have been subject to more recent introgression than those of columbi morphology, giving them a “higher than average” (i.e., more than ca 12%) ancestry from Late Pleistocene primigenius, a hypothesis that requires testing with DNA evidence.
Even the second proposed hybridisation, in the Late Pleistocene, produced ‘advanced’ mammoths of ‘jeffersonii’ type only sporadically (Lister, 2017). In the analysis by Chang et al. (Chang et al., 2017, fig. 1), of 16 North American mammoths of the Late Pleistocene, presumably therefore ‘double hybrids’, most had been morphologically classified as plain columbi (such as the ‘Nip’ individual), only two as ‘jeffersonii’. Moreover, Lister and Sher (2015, SI) found that samples of mammoth molars previously classified as ‘jeffersonii’ (such as those from Jones and Trolinger Springs, Missouri: Saunders, 1988) fall metrically within the range of Late Pleistocene mammoths referred to M. columbi (Lister, 2017, fig. 2; Figure 6 herein). Within both of these groups are individuals of more ‘primigenius- like’ morphology that could be informally termed ‘Jeffersonian’ if wished (Figure 6). Such individuals (including, for example, those described by Saunders et al., 2010) may perhaps have been subject to more recent introgression than those of columbi morphology, giving them a “higher than average” (i.e., more than ca 12%) ancestry from Late Pleistocene primigenius, a hypothesis that requires testing with DNA evidence.
Why did the introgression of 50% of the woolly mammoth genome do nothing to alter the molar morphology of Middle Pleistocene North American mammoths, if the hybrids were so successful? Perhaps primigenius genes conferred advantages in other parts of the phenotype, including those of a physiological or soft-tissue nature, or in life-history characteristics such as fecundity. Alternatively, selection may have continued to favour columbi morphology over that of primigenius in the more southerly, temperate environment of continental North America than in the northern areas where primigenius originated and persisted. It may also be that the partly intermediate, ‘jeffersonii’ morphology had nothing to do with the hybridisation at all and was due to local selection or drift. Finally, it could be that the characters on which we identify mammoth molars to these species - principally plate number - were controlled by one or a few genes, and on 50% hybridisation the ‘primigenius’ section of these could have been lost. Sections of the hybrid genome are of ‘Krestovka’ (endemic North American) origin (van der Valk et al., 2021, fig. 13 in SI) - these sections might have included the genes for plate number, purely by chance. Whatever the mechanism, there are modern parallels. Hybrid speciation in Heliconius butterflies produced a wing pattern identical to that of the parent species that contributed only 0.7% of the hybrid genome overall, and very different from that of the parent species contributing 99.3% of the hybrid genome (Frayer and Coughlan, 2024).
We have assumed until now that the first hybridisation, between the Krestovka and Chukochya lineages, happened in North America, although it could in theory have occurred in Eurasia followed by dispersal of the hybrid population into North America. The latter model would, however, require that the pre-existing North American mammoths went extinct to be replaced by the hybrid (perhaps by competition), and that the hybrid population went extinct in Siberia since no trace of Krestovka ancestry is found in any later Siberian woolly mammoths. Hybridisation in North America is therefore more plausible, but this brings its own palaeontological puzzle. While primigenius-type mammoths are known from NE Siberia back to at least 500 ka (early Middle Pleistocene), the first hybridisation implies their entry into North America long before their known fossil record. However, while the oldest finite radiocarbon date for woolly mammoths in Alaska/Yukon is ca. 60 cal ka (Mann et al., 2013), this is the limit of the method and there are many ‘infinite’ dates on specimens that could be as old as Middle Pleistocene. If this is correct, we would predict that future dating (molecular, geochronological or biostratigraphic) of primigenius-like specimens in northern North America will include individuals dating to 400 ka or more. The continuity or extinction of this population thereafter would help decide whether the second wave came from this population or from Siberia, as discussed above.
Finally, we briefly consider possible biogeographic and population scenarios to explain the 50/50 introgression seen in Mammuthus columbi. In one, females of a small population of woolly mammoths migrated into North America and mated with endemic North American males (ultimately derived from the ‘Krestovka’ population). The offspring were then incorporated into the woolly herd, which rapidly became a hybrid herd and expanded at the expense of endemic herds. In a second scenario, hybridisation occurred at a time when the population sizes of both the endemic and the immigrating population were small. However, if the more temperate-adapted endemic mammoths ever contracted to a small single refugium it would likely have been in the mid-latitudes or south of the continent, not in the far north, and likely during a glacial period. In this case the immigrant primigenius population would have had to disperse far southward to meet them.
TAXONOMIC AND NOMENCLATURAL ISSUES
Siberian trogontherii-like Mammoths
One of the consequences of any phylogenetic analysis is the potential shaking-up of established taxonomy, with the danger that the next analysis will change the taxonomy back again, or to a third position, and so on. We should therefore adopt a conservative approach to changing taxonomy, especially at the species level where it is not just a question of how to group species into higher-level taxa but how to delineate and name species in the first place. The study of van der Valk et al. (2021) was groundbreaking in that it was the first to trace genetic lineages over a million years or more. Nonetheless it was based on single individuals from Adycha, Krestovka, Chukochya, and Wyoming, with the key Krestovka genome being relatively poorly sampled. Inferences had to be made over long timescales, with no data from North American mammoths older than ca. 50 ka. These limitations urge caution in taxonomic revision, but future work has the potential to produce older DNA from Europe and North America, as well as further early material from Siberia, to confirm and clarify the relationships among these lineages.
A significant question is whether the Adycha and Krestovka lineages of trogontherii-like mammoths in Siberia should be considered as separate species. For example, researchers have recently split the ‘golden jackal’ (Canis aureus) into two different species (golden jackal, C. aureus and golden wolf, C. lupaster) on the basis of paraphyly revealed genomically and their geographical separation between Asia and Africa (Koepfli et al., 2015; Gopalakrishnan et al., 2018). Like the Olyorian Mammuthus, the two Canis species are difficult to separate morphologically (Koepfli et al., 2015). The fact that the Adycha and Krestovka lineages are paraphyletic (with respect to M. primigenius and M. columbi) in a gene tree might suggest that they too should have different species names. On the other hand, their apparent lack of interbreeding might simply have been due to geographical separation, compatible with their being intraspecific populations or perhaps subspecies. A higher coverage Krestovka genome would be required to test this. However, an assumption of the separate existence over a long period of two mammoth lineages, not reproductively isolated, is problematic for a large mammal that was likely wide-ranging on both an individual (e.g., seasonal) and populational (dispersal) scale, and that likely went through range shifts in response to repeated glacial-interglacial cycles. Nor are there geographical barriers in northern Asia of a magnitude to have plausibly forced their isolation. It therefore seems unlikely these two lineages would never have met, implying - given the lack of genetic evidence for interbreeding - that they were reproductively isolated, and hence species under the biological species concept. Or if they were so ecologically separated that they never met, this would tend to support separate species status. However, this must be considered speculative and is apparently contradicted by the later successful hybridisation between the descendants of the Krestovka and Adycha lineages in North America.
Given this uncertainty, the decision whether to award species status could alternatively be based on degree of genomic divergence, or time since divergence, but as in all such cases this is somewhat arbitrary. The time to achieve hybrid inviability in mammals and birds has been estimated as two million years or more (Price and Bouvier, 2002; Fitzpatrick, 2004). However, DNA-based estimates of time since separation of mammalian sister-species vary greatly, and there are examples of recent speciation events within the last 0.8-0.4 myr in, for example, polar/brown bears and neanderthal/modern humans (Green et al., 2010; Liu et al., 2014). The roughly one-million-year divergence time estimate for the Adycha and Krestovka lineages is therefore consistent with full speciation. Conversely, there are examples of population divergence for similar lengths of time which are still considered intraspecific. Examples based on mtDNA include red deer, Cervus elaphus, with time to most recent common ancestor (TMRCA) of its subspecies ca. 0.73 Ma (95% range 0.52-0.95 Ma) (Meiri et al., 2018), and species of African jackal with TMRCA of ca. 1.4 Ma (Lupulella adusta) and even ca. 2.6 Ma (L. mesomelas) (Atickem et al., 2018). We cannot therefore decide the status of the Krestovka and Adycha lineages based on their genetic distance or inferred divergence time.
Many authors have stressed that species delineation should be based on “a conservative consensus across a wide range of methods” (Carstens et al., 2013) or “the maximum number of available characters, and that the ideal scenario involves a combination of ecological, behavioural, phenotypic and genotypic data” (Tobias et al., 2010). Stanton et al. (2019) emphasised adaptive, behavioural, and ecological differences, as well as degree of reproductive isolation, as criteria for dividing species, agreeing that genetic distance (or time) alone is a poor guide. Nonetheless, genetic data can contribute information on these broader criteria (e.g., those involved in adaptation or reproductive isolation). Analysis of the Adycha genome (van der Valk et al., 2021) highlighted changes in functional genes, some of them believed to be adaptive to the arctic habitat. However, none of these changes are covered in the more fragmentary Krestovka genome so we cannot assess potential adaptive differences between them. In sum, more data are needed to resolve if the Adycha and Krestovka lineages were different species, or simply divergent populations of one species. This question could be resolved, firstly, by analysing more specimens to investigate if the two lineages really did coexist in both space and time (e.g., by finding them together in the same geological locality and layer, or via molecular dating). Secondly, sequencing the entire genomes of the Krestovka and Adycha lineages would allow us to investigate how large a proportion of the genome supports monophyly (Paijmans et al., 2021).
If the Krestovka and Adycha mammoths were to be considered separate species on this basis, then currently only the two genotyped specimens are taxonomically identifiable and the hundreds of other mammoth fossils from the Early Olyorian, with no DNA evidence, could not be assigned to one or the other of these species and would have to be considered Mammuthus sp. indet. If, conversely, we retain these two lineages as conspecific with each other, all remain as M. cf. trogontherii, or more cautiously trogontherii-like, on the basis of their dental similarity to the European type material of that species and other Eurasian samples referred to it for which, without exception, we do not have any DNA data. Not until the Adycha lineage gave rise to the Chukotka lineage, that is distinct morphologically as well as on DNA, is a name change clearly warranted among Siberian mammoths, and it already has a separate species name - Mammuthus primigenius. Ultimately, we may gain morphological evidence that Adycha and Krestovka were separate species; like them, the living African elephant species Loxodonta cyclotis and L. africana are not distinguishable on teeth, apart from size (Lister, 2013) but they differ strongly in their skulls (Groves and Grubb, 2000); thus far we have no Olyorian mammoth skulls. Future research may also determine whether either of the Siberian trogontherii-like lineages is actually conspecific with the European type material of M. trogontherii.
Status of the Columbian Mammoth
The hybridisation event is a remarkable discovery, showing that Late Pleistocene mammoths in North America are derived from the Siberian Early Pleistocene Krestovka lineage, with a substantial input of genes from the primigenius lineage in the Middle Pleistocene. An interesting question is whether this hybridisation event should be considered the ‘origin’ of the Columbian mammoth Mammuthus columbi. In favour of this argument is the roughly 50/50 composition of the resulting genome, so that from a purely genetical point of view it would be hard to consider the hybrid population as equivalent to one or other of the parent species. Further, if a criterion for hybrid speciation is that the two parent populations are themselves good species, then this criterion is fulfilled, because by the Middle Pleistocene the descendants of Chukotka are defined as M. primigenius, while the endemic North American mammoths are morphologically still of the trogontherii/columbi type.
On the other hand, the ‘hybrid’ is morphologically indistinguishable, on current evidence, from its predecessors in North America (see above). In other words, its molar teeth continued to look like the Krestovka (trogontherii- like) parent, not the post-Chukotka (primigenius- like) parent or an intermediate. As discussed above, this implies selection against hybrids with a more ‘primigenius- like’ dental morphology, or that the latter was lost by chance, so the original American phenotype remained unaltered. Other features, such as mandibular morphology, have not been shown to differ significantly between the two parental forms (Lister and Sher, 2015; Lister, 2017).
Dowling and Secor (1997) discussed the difficulties of recognising hybrid speciation. They concluded that “a hybrid taxon is an independently evolving, historically stable population or group of populations possessing a unique combination of heritable characteristics derived from interbreeding of representatives from two or more discrete units (e.g., races, subspecies, species, etc.). Historical stability implies that the mosaic of characters inherited from independent lineages is retained in the population, passed on from parent to offspring”. Under this definition the mosaic genetic nature of Late Pleistocene North American Columbian mammoths, following one or more hybridisation events, would qualify it as a hybrid taxon.
However, other authors have considered that further criteria are required to demonstrate hybrid speciation: the hybrids must be reproductively isolated from the parental species, and there should be evidence that hybridization is the cause of the isolation (Mavarez et al., 2006; Schumer et al., 2014). This definition has led to considerable debate, other authors considering the isolation criteria too stringent (Feliner et al., 2017). Mallet (2007) suggested that reproductive isolation is not required but “a hybrid species [is] a hybrid form that has become stabilized and remains distinct when in contact with either parent”, i.e., even in the face of some backcrossing. It is difficult to assess these criteria with respect to Late Pleistocene Columbian mammoths. Since the product of the first hybridisation in North America later hybridised with woolly mammoths - a later and more derived descendent of one of its parent lineages - the criterion of isolation is not fulfilled in this case. Did it at least remain distinct? Some individuals, even bearing DNA evidence of the second hybridisation (e.g., the Wyoming Columbian mammoth ‘Nip’), do not show any morphological alteration, while others (‘jeffersonian’ types) do. Until we have more information on the genetics of these populations, and their relation to morphology, it is impossible to resolve these questions.
In a perspective more closely applicable to palaeontological morphospecies, Schumer et al. (2018) suggest that introgression due to hybridisation, to be considered a speciation event, must have led to morphological or ecological changes that alter the subsequent evolutionary trajectory of the lineage. They cite their study of swordtail fishes (Xiphophorus) where a hybrid lineage did not differ phenotypically in any meaningful way from one of its ancestors because selection had “purged hybrid ancestry from the most functionally important regions of the genome”. This case appears potentially similar to that of the Columbian mammoth, where there is no evident adaptive change, at least in the molars, as a result of the first hybridisation. As well as morphological stasis, analysis of functional loci (van der Valk et al., 2021) showed that the bulk of adaptive changes in the mammoth lineage, including cold-adaptive features, were already present in the Adycha lineage, with relatively little change from there to Chukochya and later woolly mammoths. As long-term Arctic residents, the same may have been true of the Krestovka lineage, or they could have been more temperate adapted, extending to the far north only in warm intervals; we currently have no evidence on this point. However, once in North America, the early ‘Krestovka’ immigrants had rapidly dispersed as far south as Florida and even Mexico, where they lived for 0.5-1.0 myr before hybridisation with incoming primigenius began, so they likely were either already adapted to more temperate conditions or readapted to them. In either case the hybridisation, when it came, was between two rather differently adapted lineages. Numerous studies in extant species have shown that, for a single trait, hybrids may be intermediate, resemble one parent or the other, or show a novel phenotype unlike either Mallet, 2007 or Soltis, 2013). In their molar traits the hybrid mammoths retained endemic North American morphology, so on the criteria of Schumer et al. (2018) the case might not be considered hybrid speciation. Across traits, however, hybrids often show a mosaic condition, and this may prove to be the case in the mammoth example: in dental morphology the hybrid resembles one parent, but in others, yet to be examined, it may differ, potentially producing a unique condition justifying its recognition as a hybrid species.
Taxonomy of North American Mammoths
The new genomic data have several important consequences for the taxonomy of North American mammoths. First, they provide no evidence in support of the contention that early (pre-hybridisation) mammoths in North America were more ‘primitive’ morphologically than Late Pleistocene M. columbi, to be equated with the Eurasian M. meridional is or a local equivalent like ‘M. hayi’. Quite to the contrary, the DNA evidence for the origin of North American mammoths from the Krestovka lineage supports the observation that the morphology that seeded N America was already of advanced, ‘trogontherioid’ type.
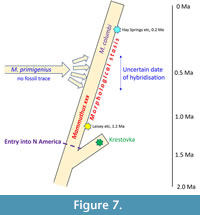
The subsequent hybridisation of these mammoths with incoming primigenius populations has differing taxonomic consequences depending on its interpretation. If we were to accept the hybridisation as a speciation event, i.e., the ‘origin of the Columbian mammoth’, this implies that only post-hybridisation North American mammoths should be called Mammuthus columbi. The type specimen of M. columbi comes from a Rancholabrean (ca. 200-12 ka: Bell et al., 2004, p. 286) locality in Georgia (Lister, 2017), and therefore presumably hosted the ‘hybrid’ genome (from the first hybridisation event at least). In this case, pre-hybridisation mammoths would require a different name, with a nominated type specimen.
However, this would pose several significant practical difficulties for researchers and curators. Since there are no clear morphological characteristics of the putative ‘early species’, nor any fossils with DNA evidence, there is no reliable way to select a type specimen on which such a taxon could be based. By the same token, the only way we could know whether a North American mammoth fossil was columbi or its predecessor would be its age - is it older or younger than the hybridisation event? But not only are many mammoth fossils poorly dated or not dated at all, but the date of the hybridisation event is itself poorly constrained, with a median estimate of 420 ka but a maximum possible range of 800-125 ka (see above).
There is therefore at present no workable solution for palaeontologists than to continue to employ a morphospecies concept and label all columbi-like mammoths as Mammuthus columbi, until and unless we find some distinguishing features that changed at the hybridisation event. In this way the hybrid is considered a continuation of the Krestovka/early North American morphospecies, albeit with genetic input from M. primigenius. And since the name-bearing type of M. columbi is a Late Pleistocene specimen, this means applying the name M. columbi to the North American lineage from its inception (Lister, 2017; Figure 7). Studies so far have focussed largely on molar teeth, and to a lesser extent on mandibles, so it may be that future studies of crania and postcrania will reveal such differences. Any such future taxonomy should be based on quantitative, statistical analysis of samples across time and space to ensure its reliability.
Late Pleistocene morphotypes not typically like either columbi or primigenius, including ‘intermediate’ forms, also need to be incorporated within the taxonomic framework. These morphotypes are very patchy even among demonstrably double-hybrid individuals. These individuals may, as suggested above, result from differing degrees of hybridisation, or may be the result of local selection. Unless shown to have a populational basis with clear chronological or geographical coherence, M. jeffersonii as a species is not a reliable concept. These individuals can be considered morphs of M. columbi, individuals of which could informally be termed ‘jeffersonian’ (Lister, 2017).
A final point of discussion concerns the taxonomic relationship between Old and New World mammoths. If the early North American mammoths were to receive a new name, then we might logically have to extend it to the Krestovka lineage of Early Olyorian mammoths in Siberia, to which they are genetically related. And if so, should this extend to the Adycha lineage as well? This would depend on the questions, yet unresolved, whether Krestovka and Adycha should be recognised as separate species, and whether either of them is conspecific with M. trogontherii of Europe (see above). Lister and Sher (2015) pointed out that European trogontherii and North American columbi are so similar that their taxonomic distinction is largely a matter of usage. If European M. trogontherii, the Siberian Krestovka lineage, and North American M. columbi were the same species, Mammuthus columbi Falconer, 1856, would technically have date priority over trogontherii Pohlig, 1885. However, the prospect of curators in every European and Asian country where M. trogontherii has been discovered renaming them to M. columbi is fanciful. Alternatively, if the species M. columbi is limited to post-hybridisation North American mammoths, and Krestovka were shown to be conspecific with European trogontherii, then this name should logically be extended to early North American mammoths as well. Lister and Sher (2015) tentatively referred Early Pleistocene molars from the Yukon Territory to M. trogontherii, from their similarity to coeval Olyorian material and also their shared biogeographic position within Beringia. To extend this terminology to all early North American mammoths is taxonomically feasible but suffers from the same boundary problems as discussed above: how to separate American trogontherii from columbi without DNA?
Our recommendation, until we demonstrate clear morphological and/or genetic differences between earlier and later North American mammoths, is to adopt a pragmatic approach that retains all North American mammoth fossils in M. columbi (aside from Late Pleistocene M. exilis and M. primigenius). If it is wished to acknowledge the DNA evidence, while remaining consistent with the morphology, we could call pre-hybridisation North American mammoths M. cf. columbi, or even just ‘columbi-like’, though as discussed above, separating them from ‘true columbi’ could only be based on rough appeal to geological age. Moreover, ‘columbi-like’ lacks formal taxonomic status.
Similarly for Eurasian trogontherii, we suggest that palaeontologists have little choice for now but to continue using the accepted morphospecies. European Middle Pleistocene fossils are M. trogontherii, and closely similar Chinese and NE Siberian Early Pleistocene ones can be referred to M. cf. trogontherii, or in the absence of DNA evidence to link them to the European form we can descriptively refer to them as ‘trogontherii-like’. Similar issues concern the status of some island mammoth populations, for example, Japanese fossils similar to M. trogontherii that have been separated as a related form, Mammuthus protomammonteus (Matsumoto, 1924) on morphological grounds (Takahashi and Namatsu, 2000; Figure 2). Future genomic work will hopefully shed light on these issues.
CONCLUSIONS
Recent genomic evidence has paved the way for further, exciting research into mammoth evolution. A more complete genome from the Krestovka lineage is required, as well as overcoming the technical challenges of obtaining DNA from Early to Middle Pleistocene deposits in the more southerly regions of Eurasia and North America. Other burgeoning tools, such as stable isotope and proteomic analysis, will likely complement genomic data in furthering insights into relationships and adaptation. On the morphological side, we need to look beyond molar teeth to clues from the detailed anatomy of skulls and mandibles, with their own challenges of overcoming substantial sexual and ontogenetic variation in these structures (Lister, 2017, fig. S3).
ACKNOWLEDGEMENTS
The authors are grateful to the referees of this paper for their helpful suggestions. L. Dalén acknowledges support from the Swedish Research Council (2021-00625) and the European Union (ERC, PrimiGenomes, 101054984).
REFERENCES
Agenbroad, L.D. 2005. North American proboscideans: mammoths: the state of knowledge, 2003. Quaternary International, 126–128:73–92.
https://doi.org/10.1016/j.quaint.2004.04.016
Albayrak, E. and Lister, A.M. 2011. Dental remains of fossil elephants from Turkey. Quaternary International, 276–277:198–211.
https://doi.org/10.1016/j.quaint.2011.05.042
Atickem, A., Stenseth, N.C., Drouilly, M., Bock, S., Roos, C., and Zinner, D. 2018. Deep divergence among mitochondrial lineages in African jackals. Zoologica Scripta, 47:1–8.
https://doi.org/10.1111/zsc.12257
Bell, C.J., Lundelius, E.L. Jr., Barnosky, A.D., Graham, R.W., Lindsay, E.H., Ruez, D.R. Jr., Semken, H.A. Jr., Webb, S.D., and Zakrzewski, R.J. 2004. The Blancan, Irvingtonian, and Rancholabrean Mammal Agesin, p. 232–314. In Woodburne, M.O. (ed.), Late Cretaceous and Cenozoic Mammals of North America: Biostratigraphy and Geochronology. Columbia University Press, New York.
https://doi.org/10.7312/wood13040
Carstens, B.C., Pelletier, T.A., Reid, N.M., and Satler, J.D. 2013. How to fail at species delimitation. Molecular Ecology, 22:4369–4383.
https://doi.org/10.1111/mec.12413
Chang, D., Knapp, M., Enk, J., Lippold, S., Kircher, M., Lister, A., MacPhee, R.D.E., Widga, C., Czechowski, P., Sommer, R., Hodges, E., Stümpel, N., Barnes, I., Dalén, L., Derevianko, A., Germonpré, M., Hillenbrand-Voiculescu, A., Constantit, S., Kuznetsova, T., Mol, D., Rathgeber, T., Rosendahl, W., Tikhonov, A.N., Willerslev, E., Hannon, G., Lalueza-Fox, C., Joger, U., Poinar, H., Hofreiter, M., and Shapiro, B. 2017. The evolutionary and phylogeographic history of woolly mammoths: a comprehensive mitogenomic analysis. Scientific Reports, 7:44585.
https://doi.org/10.1038/srep44585
Dowling, T.E. and Secor, C.L. 1997. The role of hybridisation and introgression in the diversification of animals. Annual Review of Ecology and Systematics, 28:593–619.
https://doi.org/10.1146/annurev.ecolsys.28.1.593
Enk, J., Devault, A., Debruyne, R., King, C.E., Treangen, T., O’Rourke, D., Salzberg, S.L., Fisher, D., MacPhee, R., and Poinar, H. 2011. Complete Columbian mammoth mitogenome suggests interbreeding with woolly mammoths. Genome Biology, 12:R51.
https://doi.org/10.1186/gb-2011-12-5-r51
Enk, J., Devault, A., Widga, C., Saunders, J., Szpak, P., Southon, J., Rouillard, J.-M., Shapiro, B., Golding, G.B., Zazula, G., Froese, D., Fisher, D.C., MacPhee, R.D.E., and Poinar, H. 2016. Mammuthus population dynamics in Late Pleistocene North America: divergence, phylogeography, and introgression. Frontiers in Ecology and Evolution, 4:42.
https://doi.org/10.3389/fevo.2016.00042
Feliner, G.N., Álvarez, I., Fuertes-Aguilar, J., Heuertz, M., Marques, I., Moharrek, F., Piñeiro, R., Riina, R., Rosselló, J.A., Soltis, P.S., and Villa-Machío, I. 2017. Is homoploid hybrid speciation that rare? An empiricist’s view. Heredity, 118:513–516.
https://doi.org/10.1038/hdy.2017.7
Fitzpatrick, B.M. 2004. Rates of evolution of hybrid inviability in birds and mammals. Evolution, 58:1865–1870.
https://doi.org/10.1111/j.0014-3820.2004.tb00471.x
Frayer, M.E. and Coughlan, J.M. 2024. Surprise hybrid origins of a butterfly species. Nature, 628:723–724.
https://doi.org/10.1038/d41586-024-00858-3
Gopalakrishnan, S., Sinding, M.-H.S., Ramos-Madrigal, J., Niemann, J., Samaniego Castruita, J.-A., Vieira, F.G., Carøe, C., Manuel Montero, M. de, Kuderna, L., Serres, A., González-Basallote, V.M., Liu, Y.-H., Wang, G.-D., Marques-Bonet, T., Mirarab, S., Fernandes, C., Gaubert, P., Koepfli, K.-P., Budd, J., Rueness, E.K., Sillero, C., Heide-Jørgensen, M.P., Petersen, B., Sicheritz-Ponten, T., Bachmann, L., Wiig, Ø., Hansen, A.J., and Gilbert, M.T.P. 2018. Interspecific Gene Flow Shaped the Evolution of the Genus Canis. Current Biology, 28:3441–3449.
https://doi.org/10.1016/j.cub.2019.11.009
Green, R.E., Krause, J., Briggs, A.W., Maricic, T., Stenzel, U., Kircher, M., Patterson, N., Li, H., Zhai, W., Fritz, M.H.Y., Hansen, N.F., Durand, E.Y., Malaspinas, A.-S., Jensen, J.D., Marques-Bonet, T., Alcan, C., Prüfer, K., Meyer, M., Burbano, H.A., Good, J.M., Schultz, R., Aximu-Petri, A., Butthof, A., Höber, B. Höffner, B., Siegemund, M., Weihmann, A., Nusbaum, C., Lander, E.S., Russ, C., Novod, N., Affourtit, J., Egholm, M., Verna, C., Rudan, P., Brajkovic, D., Kucan, Ž., Gušic, I., Doronichev, V.B., Golovanova, L.V., Lalueza-Fox, C., de la Rasilla, M., Fortea, J., Rosas, A., Schmitz, R.W., Johnson, P.L.F., Eichler, E.E., Falush, D., Birney, E., Mullikin, J.C., Slatkin, M., Nielsen, R., Kelso, J., Lachmann, M., Reich, D., and Pääbo, S. 2010. A draft sequence of the Neandertal genome. Science, 328:710–722.
https://doi.org/10.1126/science.1188021
Groves, C.P. and Grubb, P. 2000. Do Loxodonta cyclotis and L. africana interbreed? Elephant, 2: 4–7.
https://doi.org/10.22237/elephant/1521732173
Harington, C.R. 1984. Mammoths, bison and time in North America, p. 299–309. In Mahaney, W.C. (ed.), Quaternary Dating Methods. Elsevier, Amsterdam.
https://doi.org/10.1016/S0920-5446(08)70078-5
Koepfli, K.P., Pollinger, J., Godinho, R., Robinson, J., Lea, A., Hendricks, S., Schweizer, R.M., Thalmann, O., Silva, P., and Fan, Z. 2015. Genome-wide Evidence Reveals that African and Eurasian Golden Jackals Are Distinct Species. Current Biology, 25:2158–2165.
https://doi.org/10.1016/j.cub.2015.06.060
Lister, A.M. 1996. Evolution and taxonomy of Eurasian mammoths, p. 203–213. In Shoshani, J. and Tassy, P. (eds.), The Proboscidea: Trends in Evolution and Paleoecology. Oxford University Press.
https://doi.org/10.1093/oso/9780198546528.001.0001
Lister, A.M. 2013. Behavioural leads in evolution - evidence from the fossil record. Biological Journal of the Linnean Society, 112:315–331.
https://doi.org/10.1111/bij.12173
Lister, A.M. 2017. On the type material and evolution of North American mammoths. Quaternary International, 443A:14–31.
https://doi.org/10.1016/j.quaint.2017.02.027
Lister, A.M. 2022. Mammoth evolution in the late Middle Pleistocene: The Mammuthus trogontherii-primigenius transition in Europe. Quaternary Science Reviews, 294:107693.
https://doi.org/10.1016/j.quascirev.2022.107693
Lister, A. and Bahn, P. 2007. Mammoths: Giants of the Ice Age. University of California Press.
Lister, A.M. and Sher, A.V. 2001.The origin and evolution of the woolly mammoth. Science, 294:1094–1097.
https://doi.org/10.1126/science.1056370
Lister, A.M. and Sher, A.V. 2015. Evolution and dispersal of mammoths across the Northern Hemisphere. Science, 350:805–809.
https://doi.org/10.1126/science.aac5660
Liu, S., Lorenzen, E.D., Fumagalli, M., Li, B., Harris, K., Xiong, Z., Zhou, L., Sand Korneliussen, T., Somel, M., Babbitt, C., Wray, G., Li, J., He, W., Wang, Z., Fu, W., Xiang, X., Morgan, C.C., Doherty, A., O’Connell, M.J., McInerney, J.O., Born, E.W., Dalén, L., Dietz, R., Orlando, L., Sonne, C., Zhang, G., Nielsen, R., Willerslev, E., and Wang, J. 2014. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell, 157:785–794.
https://doi.org/10.1016/j.cell.2014.03.054
Maglio, V.J. 1973. Origin and Evolution of the Elephantidae. Transactions of the American Philosophical Society, New Series, 63:1–149.
https://doi.org/10.2307/1006229
Mallet, J. 2007. Hybrid speciation. Nature, 446:279–283.
https://doi.org/10.1038/nature05706
Mann, D.H., Groves, P., Kunz, M.L., Reanier, R.E., and Gaglioti, B.V. 2013. Ice-age megafauna in Arctic Alaska: extinction, invasion, survival. Quaternary Science Reviews, 70:91–108.
https://doi.org/10.1016/j.quascirev.2013.03.015
Mavarez, J., Salazar, C.A., Bermingham, E., Salcedo, C., Jiggins, C.D., and Linares, M. 2006. Speciation by hybridisation in Heliconius butterflies. Nature, 441:868–871.
https://doi.org/10.1038/nature04738
McDaniel, G.E. and Jefferson, G.T. 2003. Mammuthus meridionalis (Proboscidea: Elephantidae) from the Borrego Badlands of Anza-Borrego Desert State Park, California: phylogenetic and biochronologic implications. Deinsea, 9:239–252.
Meiri, M., Kosintsev, P., Conroy, K., Meiri, S., Barnes, I., and Lister, A.M. 2018. Subspecies dynamics in space and time: a study of the red deer complex using ancient and modern DNA and morphology. Journal of Biogeography, 45:367–380.
https://doi.org/10.1111/jbi.13124
Osborn, H.F. 1942. Proboscidea. Volume 2. American Museum of Natural History, New York.
Paijmans, J.L.A., Barlow, A., Becker, M.S., Cahill, J.A., Fickel, J., Förster, D.W.G., Gries, K., Hartmann, S., Havmøller, R.W., Henneberger, K., Kern, C., Kitchener, A.C., Lorenzen, E.D., Mayer, F., and O’Brien, S.J. 2021. African and Asian leopards are highly differentiated at the genomic level, Current Biology, 31:1–11.
https://doi.org/10.1016/j.cub.2021.03.084
Palkopoulou, E., Lipson, M., Mallick, S., Nielsen, S., Rohland, N., Baleka, S., Karpinski, E., Ivanevic, A.M., To, T.-H., Kortschak, R.D., Raison, J.M., Qu, Z., Chin, T.-J., Alt, K.W., Claesson, S., Dalén, L., MacPhee, R.D.E., Meller, H., Roca, A.L., Ryder, O.A., Heiman, D., Young, S., Breen, M., Williams, C., Aken, B.L., Ruffier, M., Karlsson, E., Johnson, J., Di Palma, F., Alfoldi, J., Adelson, D.L., Mailund, T., Munch, K., Lindblad-Toh, K., Hofreiter, M., Poinar, H., and Reich, D. 2018. Genomic history of extinct and living elephantids. Proceedings of the National Academy of Sciences, 115:E2566–E2574.
https://doi.org/10.1073/pnas.1720554115
Price, T.D. and Bouvier, M.M. 2002. The evolution of F1 postzygotic incompatibilities in birds. Evolution, 56:2083–2089.
https://doi.org/10.1111/j.0014-3820.2002.tb00133.x
Roca, A.L. 2021. A mammoth step back in genomic time. Nature, 591:208–209.
https://doi.org/10.1038/d41586-021-00348-w
Rohland, N., Reich, D., Mallick, S., Meyer, M., Green, R.E., Georgiadis, N.J., Roca, A.L., and Hofreiter, M. 2010. Genomic DNA sequences from mastodon and woolly mammoth reveal deep speciation of forest and savanna elephants. PLoS Biology, 8:e1000564.
https://doi.org/10.1371/journal.pbio.1000564
Saunders, J.J. 1988. Fossiliferous spring sites in southwestern Missouri. Bulletin of the Buffalo Society of Natural Science, 33:127–149.
Saunders, J.J., Grimm, E.C., Widga, C.C., Campbell, G.D., Curry, B.B., Grimley, D.A., Hanson, P.R., McCullum, J.P., Oliver, J.S., and Treworgy, J.D. 2010. Paradigms and proboscideans in the southern Great Lakes region, USA. Quaternary International, 217:175–187.
https://doi.org/10.1016/j.quaint.2009.07.031
Schumer, M., Rosenthal, G.G., and Andolfatto, P. 2014. How common is homoploid hybrid speciation? Evolution, 68:1553–1560.
https://doi.org/10.1111/evo.12399
Schumer, M., Rosenthal, G.G., and Andolfatto, P. 2018. What do we mean when we talk about hybrid speciation? Heredity, 120:379–382.
https://doi.org/10.1038/s41437-017-0036-z
Sher, A.V. 1986. Olyorian land mammal age of northeastern Siberia. Palaeontographia Italica, 74:97–112.
Soltis, P.S. 2013. Hybridisation in plants, p. 166–176. In Levin, S.A. (ed.), Encyclopaedia of Biodiversity. Elsevier, Amsterdam.
Stanton, D.W.G., Frandsen, P., Waples, R.K., Heller, R., Russo, I.-R., Orozco-terWengel, P.A., Pedersen, C.-E. T., Slegismund, H.R., and Bruford, M.W. 2019. More grist for the mill? Species delimitation in the genomic era and its implications for conservation. Conservation Genetics, 20:101–113.
https://doi.org/10.1007/s10592-019-01149-5
Sutcliffe, A.J. 1985. On the Track of Ice Age Mammals. British Museum (Natural History), London.
Takahashi, K. and Namatsu, K. 2000. Origin of the Japanese Proboscidea in the Plio-Pleistocene. Earth Science (Chikyu Kagaku), 54:257–267.
https://doi.org/10.15080/agcjchikyukagaku.54.4_257
Tobias, J.A., Seddon, N., Spottiswoode, C.N., Pilgrim, J.D., Fishpool, L.D.C., and Collar, N.J. 2010. Quantitative criteria for species delimitation. Ibis, 152:724–746.
https://doi.org/10.1111/j.1474-919X.2010.01051.x
van der Valk, T., Pečnerová, P., Díez-del-Molino, D., Bergström, A., Oppenheimer, J., Hartmann, S., Xenikoudakis, G., Thomas, J.A., Dehasque, M., Sağlıcan, E., Fidan, F.R., Barnes, I., Liu, S., Somel, M., Heintzman, P.D., Nikolskiy, P., Shapiro, B., Skoglund, P., Hofreiter, M., Lister, A.M., Götherström, A., and Dalén, L. 2021. Million-year-old DNA sheds light on the genomic history of mammoths. Nature, 591:265–269.
https://doi.org/10.1038/s41586-021-03224-9
Wei, G., Taruno, H., Kawamura, Y., and Jin, C. 2006. Pliocene and Early Pleistocene primitive mammoths of Northern China: Their revised taxonomy, biostratigraphy and evolution. Journal of Geosciences Osaka City University, 49:59–101
Wei, G., Hu, S., Yu, K., Hou, Y., Jin, C., Wang, Y., Zhao, J., and Wang, W. 2010. New materials of the steppe mammoth, Mammuthus trogontherii, with discussion on the origin and evolutionary patterns of mammoths. Science China Earth Sciences, 53:956–963.
https://doi.org/10.1007/s11430-010-4001-4

