 X-ray micro-CT reconstruction reveals eight antennomeres in a new fossil taxon that constitutes a sister clade to Dundoxenos and Triozocera (Strepsiptera: Corioxenidae)
X-ray micro-CT reconstruction reveals eight antennomeres in a new fossil taxon that constitutes a sister clade to Dundoxenos and Triozocera (Strepsiptera: Corioxenidae)
Article number: 16.3.29A
https://doi.org/10.26879/370
Copyright Palaeontological Association, December 2013
Author biographies
Plain-language and multi-lingual abstracts
PDF version
Submission: 7 January 2013. Acceptance: 5 November 2013
{flike id=552}
ABSTRACT
Eocenoxenos palintropos gen. nov. et sp.nov., a new fossil strepsipteran taxon from Baltic amber is described. The position of the new genus is based on cladistic analyses of morphological data sets. Most data of the fossil where retrieved with 3D micro-CT scan reconstructions. The new taxon is unambiguously situated as a sister group of the Dundoxenos-Triozocera clade within the Corioxenidae. The eocene taxon combines derived characteristics typical of Corioxenidae with the posession of eight antennomeres with five long flabella, a regained ancestral characteristic.
Hans Henderickx. Department of Biology, Universiteit Antwerpen, Groenenborgerlaan 171, 2020 Antwerpen, Belgium (Address for correspondence: Hemelrijkstraat 4, B-2400 Mol, cavexplorer@gmail.com
Jan Bosselaers. Section of invertebrates, Royal Museum for Central Africa, B-3080 Tervuren, Belgium dochterland@telenet.be
Elin Pauwels. Department of Physics and Astronomy, Gent University, Proeftuinstraat 86, B-9000 Gent, Belgium elin.pauwels@UGent.be
Luc Van Hoorebeke. Department of Physics and Astronomy, Gent University, Proeftuinstraat 86, B-9000 Gent, Belgium luc.vanhoorebeke@UGent.be
Matthieu Boone. Department of Physics and Astronomy, Gent University, Proeftuinstraat 86, B-9000 Gent, Belgium matthieu.boone@UGent.be
KEY WORDS: Strepsiptera; new genus; new species; micro-CT scan; Baltic amber fossil
Final citation: Henderickx, Hans, Bosselaers, Jan, Pauwels, Elin, Van Hoorebeke, Luc, and Boone, Matthieu. 2013. X-ray micro-CT reconstruction reveals eight antennomeres in a new fossil taxon that constitutes a sister clade to Dundoxenos and Triozocera (Strepsiptera: Corioxenidae), Palaeontologia Electronica Vol. 16, Issue 3; 29A; 16p. https://doi.org/10.26879/370
palaeo-electronica.org/content/2013/552-a-new-strepsiptera-genus
http://zoobank.org/0FAD22E6-6384-4682-A375-F59CF4BD9387
INTRODUCTION
Strepsiptera are regularly reported from Baltic amber. The fossil tree resin functioned in a way similar to a trunk eclector trap (Dubois and LaPolla, 1999) often capturing invertebrates that are seldom encountered in the field, for example because they are rare or have very short active adult live spans. Most of these Strepsiptera fossils are closely related to representatives of extant genera or species (Pohl et al., 2005; Kathirithamby and Henderickx, 2008). However, Eocene Baltic amber also contains some typical ancestral forms: the extinct genus Mengea Grote, 1886 is only found in Baltic amber and in 2005 Protoxenos janzeni Pohl et al. was described, wich has a very basal position in the group.
The new Baltic amber fossil examined here was purchased on an auction on eBay from an amber dealer in Lithuania in March 2011. It showed a puzzling combination of characteristics and could not be placed in any of the recognized genera. Its phylogenetic position is evaluated cladistically, and a new species and new genus is described.
MATERIALS AND METHODS
Amber Fossil
 The examined strepsipteran is fossilized in a 24.9 x 20.4 x 10.3 mm piece of Baltic amber (Figure 1.1). The specimen is mostly intact, and the left hind wing is spread. Larger parts of the body are covered with opaque whitish amber, 'Baltic mould' (Figure 1.1, Figure 2.1), which obscures especially the ventral side. The inclusion reaches the surface of the amber matrix with a tip of the hind wing, some amber crazing (cracks and degeneration) and some brown discoloration appear near this tip, and before further manipulation the surface was stabilized with a coating of 0.5 mm ARALDITE 2020 epoxy, using the technique described in Henderickx et al., 2006, which allows further optical observation without repolishing. Optically visible details were observed and measured using reflected and translucent illumination on a Leitz microscope and a Canon MP-E objective in combination with Zerene stacker image processing software.
The examined strepsipteran is fossilized in a 24.9 x 20.4 x 10.3 mm piece of Baltic amber (Figure 1.1). The specimen is mostly intact, and the left hind wing is spread. Larger parts of the body are covered with opaque whitish amber, 'Baltic mould' (Figure 1.1, Figure 2.1), which obscures especially the ventral side. The inclusion reaches the surface of the amber matrix with a tip of the hind wing, some amber crazing (cracks and degeneration) and some brown discoloration appear near this tip, and before further manipulation the surface was stabilized with a coating of 0.5 mm ARALDITE 2020 epoxy, using the technique described in Henderickx et al., 2006, which allows further optical observation without repolishing. Optically visible details were observed and measured using reflected and translucent illumination on a Leitz microscope and a Canon MP-E objective in combination with Zerene stacker image processing software.
X-ray Scanning and Reconstruction
The amber sample was scanned at the 'Centre for X-ray Tomography' of the Gent University (UGCT; www.ugct.ugent.be) (Masschaele et al, 2007). UGCT develops its own micro- and nano-CT scanners which are characterized by a modular structure. In this way optimal scanning parameters, such as the detector, X-ray tube, and filter materials, can be selected for each sample. For this sample the transmission head of a dual head X-ray tube from Feinfocus (FXE 160) was chosen as a source and an a:Si flat panel (PerkinElmer XRD 1620 CN3 CS) with CsI scintillator was used as detector. The X-ray tube was operated at 100kV.  The voxel size achieved for this sample was 3.0µm. Octopus (www.octopusreconstruction.com) (Vlassenbroeck et al, 2007), an in-house developed software package for parallel, fan, cone and helical cone beam geometry, was used for tomographic reconstruction. VGStudio MAX (www.volumegraphics.com) was used to render the 3D-volumes and to create an STL file. Based on this file, spatial 3D models could be created. Thomas Pilkington (Fablab, University Leuven) printed a white Acrylonitrile butadiene styrene (ABS) 3D model with fused depositioning modeling (10 cm large, layers of 0.254 mm) (Figure 2.2). Bart Grimonprez (Hogeschool West-Vlaanderen, Industrial Design Center) printed a black 3D model with polyjet modelling (5,5 cm large, layers of 0.028 mm, VeroBlack Plus). These prints appeared very useful in interpretation and observation of morphological details, such as the counting of the ommatidia and the observation of the flagellomeres.
The voxel size achieved for this sample was 3.0µm. Octopus (www.octopusreconstruction.com) (Vlassenbroeck et al, 2007), an in-house developed software package for parallel, fan, cone and helical cone beam geometry, was used for tomographic reconstruction. VGStudio MAX (www.volumegraphics.com) was used to render the 3D-volumes and to create an STL file. Based on this file, spatial 3D models could be created. Thomas Pilkington (Fablab, University Leuven) printed a white Acrylonitrile butadiene styrene (ABS) 3D model with fused depositioning modeling (10 cm large, layers of 0.254 mm) (Figure 2.2). Bart Grimonprez (Hogeschool West-Vlaanderen, Industrial Design Center) printed a black 3D model with polyjet modelling (5,5 cm large, layers of 0.028 mm, VeroBlack Plus). These prints appeared very useful in interpretation and observation of morphological details, such as the counting of the ommatidia and the observation of the flagellomeres.
Phylogenetic Analysis
Phylogenetic analyses were based on all 189 characters from the impressive data matrix of Pohl and Beutel (2005) and were performed using the computer programmes PAUP 4.0 beta 10 (Swofford, 2002), Winclada 1.00.08 (Nixon, 2002), TNT 1.1 (Goloboff et al., 2003, 2008b) and TreeRot v3 (Sorenson and Franzosa, 2007). Optimisation of character states and printing of the preferred tree was performed using Winclada, further layout modifications being executed in Adobe Photoshop Elements v8.0. All four cladistic programmes were run on a dual-core Intel iMac under a Windows XP virtual machine (VMware Fusion 2, VMware, Inc., 3401 Hillview Avenue, Palo Alto, CA 94304, USA; Bugnion et al., 2000).
Abbreviations: ACCTRAN, accelerated transformation; ci, consistency index; ci-ex, consistency index excluding parsimony-uninformative characters; DELTRAN, delayed transformation; hi, homoplasy index; hi-ex, homoplasy index excluding parsimony-uninformative characters; rc, rescaled consistency index; ri, retention index; UNAMB, unambiguous optimisation.
SYSTEMATICS
Systematic Paleontology
Family Corioxenidae Kinzelbach, 1970
The new species belongs in the Corioxenidae based on the absence of mandibles, the reduced posterior part of the labium, the elongated abdominal segment IX, and the long penis (formerly called aedeagus, but see Hünefeld et al. (2011: 299) and Pohl et al. (2012: 81)), scored as character states 33:1, 49:3, 76:1, and 79:1 in the cladistic analysis discussed in the next part of the manuscript.
New genus: type species
Eocenoxenos Henderickx and Bosselaers, gen. nov.
http://zoobank.org/28E3CEDC-86D9-4EEB-B5CE-FD4094C48AAD
Type specimen: Eocenoxenos palintropos Henderickx and Bosselaers sp. nov.
Diagnosis
The males of the new genus can be recognized by the following unique combination of characters: Eight antennomeres (Figure 2.6-7) (character state 19:1), antennomeres III-VII being flabellate, mandibles absent, five tarsomeres with no claws and a shortened, triangular tarsomere IV (67:2), abdominal segment IX ventrally elongated for reception of the elongated penis.
Eocenoxenos shares the posession of five tarsomeres (62:0) and a slender tarsomere V (68:1) with the corioxenid genera Dundoxenos and Triozocera (Triozocerinae as defined by Kathirithamby (1989)) and Proceroxenos and Uniclavus (here considered the Uniclavinae of Kathirithamby (1989)). Eocenoxenos also differs from the genera in Triozocerinae and Uniclavinae by the absence of claws (63:3, a character state shared with all other Stylopidia) and by short antennomeres V, similar in length to antennomeres III (28:0), a character state also found in the corioxenid genera Blissoxenos and Floridoxenos, considered here subfamily Blissoxeninae as described by Miyamoto and Kifune (1984).
Etymology
The new genus-group name refers to the era in which Baltic amber was formed, the Eocene. The Appendix refers to the Greek 'ξ ε ν o ζ', guest, stranger, commonly used in generic names of Strepsiptera.
New species
Eocenoxenos palintropos Henderickx and Bosselaers sp. nov.
(Figure 1, Figure 2, Figure 3, Figure 4, and Figure 5)
http://zoobank.org/60F99337-16CE-440C-9E26-8318F5308EAD
 Specimen provenance and deposition
Specimen provenance and deposition
The male holotype in Baltic amber was purchased in an auction on eBay in March 2011 by the first author and consecutively loaned to the RBIN (Royal Belgian Instistute of Natural Sciences – Entomology, Brussels) where it is available for study under registration number IG. 32.287 with the enlarged 3D prints, the microtomographic data, and the original and segmented slices.
Diagnosis
As for the genus (see above).
Description
Male holotype. Habitus Figure 5.1, body length 4.00 mm (all further measurements in mm), body with antenna 4.90; width of head 1.19; approximate length of antenna 1.00; length of hindwing 2.88; wingspan 6.64; forewing 0.88 x 0.16; length of metathorax 2.16. 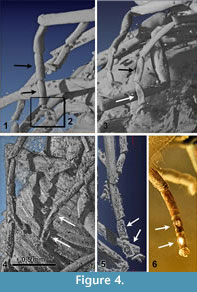 Colour very dark where not covered with whitish amber, antenna with white points (possibly chemoreceptors), ocelli bright with black interspace, however the color might be an effect of the fossilisation in amber. Head transverse, wide (Figure 5.2). Compound eye distinct, slightly oval in lateral view, right eye with 119, left eye with 122 ommatidia. Antenna 0.8 times as long as maximum width of head. Antennomeres 3-7 short, decreasing in width, Eight antennomeres, 5 flabellate antennomeres (flagellomeres I-V on antennomeres III-VII). Flabella long, flattened, widened medially, pointed. Flabella I-V all 4.3 times as long as maximum width, the flattened antennomere VIII slightly wider, 3.5 times as long as maximum width.
Colour very dark where not covered with whitish amber, antenna with white points (possibly chemoreceptors), ocelli bright with black interspace, however the color might be an effect of the fossilisation in amber. Head transverse, wide (Figure 5.2). Compound eye distinct, slightly oval in lateral view, right eye with 119, left eye with 122 ommatidia. Antenna 0.8 times as long as maximum width of head. Antennomeres 3-7 short, decreasing in width, Eight antennomeres, 5 flabellate antennomeres (flagellomeres I-V on antennomeres III-VII). Flabella long, flattened, widened medially, pointed. Flabella I-V all 4.3 times as long as maximum width, the flattened antennomere VIII slightly wider, 3.5 times as long as maximum width.
All 5 flabella approximately the same length, 0.40 mm.
Maxilla 2.1 times as long as wide, widening distally, palp1.3 times longer than maxilla, widening medially, pointed (Figure 5.2). Mandibles absent.
Venation hind wing (Figure 5.1) with vein R2 not present, R3 and R5 detached, R3 strongly developed. R5 long and running parallel to R4. CuA1 slightly shorter than CuA2.
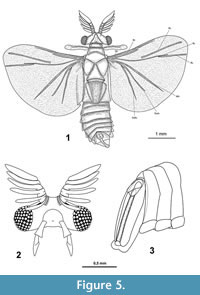 Abdominal ninth segment (genital capsule) slender, segment X folded inwards on both sides, holding the tip of the long straight penis (Figure 5.3).
Abdominal ninth segment (genital capsule) slender, segment X folded inwards on both sides, holding the tip of the long straight penis (Figure 5.3).
Legs (Figure 2.3-5,Figure 4.1-6). Pro- and mesothoracic legs with trochanter and femur fused. Distal margin of tibiae straight. 5 tarsomeres without claws. Tarsomere 2 and 3 of all legs with an oval latero-ventral sensory spot that is dented in lateral view (compared with Pohl and Beutel, 2004). Basitarsus of prothoracic legs cylindrical, tarsomere 4 shortened and triangular, diameter of tarsomere 5 less than 50% of tarsomere 4. Metacoxa immobilised and integrated into metathorax.
Etymology
The specific epithet is derived from the classical Greek π σ λ ι ν τ ρ o π o ζ, returning, reverting. It refers to the fact that the present species seems to have regained eight antennomeres, a characteristic that normally belongs only in the more basal Strepsiptera.
CLADISTIC ANALYSIS
The phylogeny of Strepsiptera has been extensively studied by Pohl and Beutel (2005). Moreover, the same data matrix was re-analysed by Bravo et al. (2009) in order to elucidate the relationships of Bahiaxenos Bravo et al., 2009. Nevertheless, a cladistic analysis has to be re-run entirely when another taxon is added. Adding a single taxon to a data matrix thoroughly influences the accuracy, robustness, and node support of the results obtained, as discussed by Kim (1998: 55-56), Bremer et al. (1999), and Poe and Swofford (1999). Indeed, modifying even a single entry in a data matrix can result in a totally different outcome of the analysis (Hovenkamp, 1999). In addition to that, repeating a cladistic analysis with a slightly modified data matrix and using a somewhat different algorithm gives a much better idea of the support and the stability of the results obtained (Hovenkamp, 2009). In view of all this, it was decided to repeat the entire analysis, using different software and settings, such as running all characters unordered and using implied weighting (Goloboff, 1993).
Taxon Choice
 The data matrix from Pohl and Beutel (2005) was used, with scores for the males of Bahiaxenos (as specified in Bravo et al., 2009: appendix) and Eocenoxenos gen. n. added. As a result, a matrix with 43 taxa is obtained, encompassing 40 genera of Strepsiptera and three outgroup taxa (Maddison et al., 1984, Watrous and Wheeler, 1981): Priacma LeConte, 1874 (Coleoptera), Sialis Latreille, 1802 (Megaloptera), and Tipula Linnaeus, 1758 (Diptera). Five strepsipteran genera in the matrix are only known from fossils and presumed extinct (indicated by † in Figure 6): Cretostylops Grimaldi and Kathirithamby, 2005 (Cretaceous), Eocenoxenos gen. n. (Eocene), Mengea Grote, 1886 (Eocene), Protelencholax Kinzelbach, 1979 (Miocene), Protoxenos Pohl et al., 2005 (Eocene). Stichotrema Hofeneder, 1910 was originally considered to encompass both Miocene and Eocene fossils (Pohl et al., 2005: table 1), and recent species. The fossil species were transferred to Palaeomyrmecolax Kulicka, 2001 by Pohl and Beutel (2005: 366) and both groups are scored in the matrix, as explained in Pohl and Beutel (2005: 370).
The data matrix from Pohl and Beutel (2005) was used, with scores for the males of Bahiaxenos (as specified in Bravo et al., 2009: appendix) and Eocenoxenos gen. n. added. As a result, a matrix with 43 taxa is obtained, encompassing 40 genera of Strepsiptera and three outgroup taxa (Maddison et al., 1984, Watrous and Wheeler, 1981): Priacma LeConte, 1874 (Coleoptera), Sialis Latreille, 1802 (Megaloptera), and Tipula Linnaeus, 1758 (Diptera). Five strepsipteran genera in the matrix are only known from fossils and presumed extinct (indicated by † in Figure 6): Cretostylops Grimaldi and Kathirithamby, 2005 (Cretaceous), Eocenoxenos gen. n. (Eocene), Mengea Grote, 1886 (Eocene), Protelencholax Kinzelbach, 1979 (Miocene), Protoxenos Pohl et al., 2005 (Eocene). Stichotrema Hofeneder, 1910 was originally considered to encompass both Miocene and Eocene fossils (Pohl et al., 2005: table 1), and recent species. The fossil species were transferred to Palaeomyrmecolax Kulicka, 2001 by Pohl and Beutel (2005: 366) and both groups are scored in the matrix, as explained in Pohl and Beutel (2005: 370).
Characters
A series of 189 characters (144 binary and 45 multistate) was coded for the 3 outgroup taxa and 40 ingroup taxa. Fourteen characters (16, 32, 46, 73, 78, 91, 96, 141, 147, 148, 155, 164, 174, and 178) are parsimony-uninformative, but have been included for the sake of completeness. Character 51 (mouthfield sclerite) is scored as binary, as character state 2 does not occur in the matrix of Pohl and Beutel (2005: table 1).
Analysis and Results
The matrix of character states, as well as the character parameters and properties on the implied weighting consensus tree, can be found in Appendix. All multistate characters were run unordered in the analyses performed, contra Pohl and Beutel (2005) and Platnick (2012). Running multistate characters as ordered, or splitting them into characters with hierarchically related character states, as advocated by Hawkins et al. (1997), did not improve the resolution or the consistency of the results obtained.
An equally weighted analysis of the data matrix was performed in PAUP with hsearch addseq = random nreps = 5000 (heuristic search with tree bisection and reconnection swapping and 5000 random addition sequences). In order to avoid spurious resolution due to unsupported (Coddington and Scharff, 1994; Wilkinson, 1995) or ambiguously supported (Nixon and Carpenter, 1996) branches, those with a minimum length of zero were collapsed with pset collapse = minbrlen. Multistate character states were coded as polymorphic with pset mstaxa = polymorph. In total, 15 shortest trees (length = 356) were found 4989 times, with a strict consensus of length 367, ci = 0.7166, ci-ex = 0.7045, hi = 0.3188, hi-ex = 0.2955, rc = 0.6219, and ri = 0.8679. The equally weighted consensus tree is identical to the tree found by Pohl and Beutel (2005, figure 28), but with Bahiaxenos branching off between Mengea and Mengenillidae, as found by Bravo et al. (2009, figure 6), and Eocenoxenos added to the unresolved clade of Dundoxenos, Triozocera, Proceroxenos, and Uniclavus.
Because cladograms obtained by attributing a posteriori weights to characters based on their relative degrees of homoplasy on a set of heuristic trees, explain the data better (Bosselaers and Jocqué, 2002; Goloboff et al., 2008a), a weighted analysis was also performed.
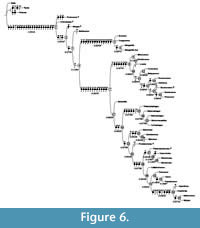
Implied weighting (Goloboff, 1993) was preferred for this purpose. When applying implied weighting in PAUP with pset goloboff = yes, pset collapse = minbrlen, pset mstaxa = polymorph, and hsearch addseq = random nreps = 1000, using a relatively mild concavity constant = 4, two fittest trees with fit = -160.33718 (PAUP attributes a negative sign to fit values) were found 218 times. The two fittest trees only differ in the position of Protoxenos and Cretostylops, which are interchanged. An additional implied weighting search run in TNT using Settings / implied weighting and Analyze / Traditional search (TBR or SPR, 1000 replications) as well as New Technology search (Collapse trees after search; Driven search, 500 init. addseqs, find min. length 100 times, 1 random seed; Sectorial search with RSS, CSS and XSS, fuse trees 10 times; Ratchet with 10 iterations; Tree drifting with 20 cycles; Tree fusing with 10 rounds), using the same concavity constant, which equals 5 instead of 4 in TNT (see Bosselaers and Jocqué, 2002: 16), produced the same two fittest trees. Because TNT calculates weighted homoplasy (a complement of fit) for implied weighting trees, another fit value, 13.33730 was reported for these same trees. The strict consensus of these two trees, which has length = 357 and fit -160.17051, PAUP value (13.50397 TNT value), is our preferred solution. It is illustrated in Figure 6. This tree has ci = 0.7367, ci-ex = 0.7251, hi = 0.2997, hi-ex = 0.2749, rc = 0.6487, and ri = 0.8806.
Bremer support (a statistic based on the number of extra steps needed on a tree in order to collapse a branch, Bremer, 1988, 1994), expressed as TNT fit values, was calculated in TNT, using Analyze / suboptimal, followed by Analyze / Traditional search / tree bisection reconnection (TBR)) and Trees / Bremer Supports, retaining trees suboptimal up to 10 units of fit and combining various numbers of replications (between 1 and 90000) with various numbers of trees saved per replication (between 1 and 90000, inversely related to the number of replications) until the solution stabilised. Bremer support was also assessed using PAUP in combination with TreeRot v3 (Sorenson and Franzosa, 2007), a programme calculating branch support using reverse constraints. For that purpose, a Nexus treefile of the implied weighting consensus tree was converted to a PAUP command file by TreeRot. This command file was subsequently run in PAUP to produce a ".results" file. Bremer support in PAUP fit values was calculated manually from the TreeRot ".results" file. Although PAUP and TNT assign different values and signs to the overall fit value of trees, Bremer support values obtained with both programmes were quite similar. In the case of small differences, the lowest (TreeRot) value was chosen.
Discussion
The preferred strict consensus tree with node numbers, state changes for 60 characters and Goloboff fit Bremer support values (as reported in TNT and PAUP) in italics below branches is illustrated in Figure 6. Each ambiguity on the tree was optimized in isolation, in order to avoid scoring character states for absent structures, and also because only a combination of ACCTRAN ("fast") and DELTRAN ("slow"" optimisation can produce the most robust proposal for a supposed homology. Indeed, the use of ACCTRAN only, as is often preferred, does not always maximize parallel loss of complex traits over convergent gains (Agnarsson and Miller, 2008). Characters 19, 29, 49, 142, 149, 172, and 177 were optimised under ACCTRAN on the tree, characters 62, 80, 92, 99, 110, 113, 119, 122,123, 128,129, and 152 were optimised under DELTRAN, and only unambiguous state changes were plotted for character 68, in order to avoid scoring this character for an absent structure, as five tarsomeres are only present in clade 13 within Stylopidia (Figure 6, clade 7; Figure 7.3).
Homoplasy is rather low in the data matrix under study: 128 out of 189 characters are completely free of homoplasy on the consensus tree (Appendix). Sanderson and Donoghue (1989: 1785, Figure 1) performed a polynomial regression analysis on data from 60 cladistic analyses, and derived the following equation based on them: ci = 0.90 - 0.022*(number of taxa) + 0.000213*(number of taxa)2. Applying this equation, 43 taxa would yield a ci value of 0.3478. The preferred tree has a significantly higher ci value of 0.7251, excluding parsimony-uninformative characters.
The consensus tree obtained (Figure 6) is almost identical to the tree of Pohl and Beutel (2005, figure 28), the only differences being Bahiaxenos branching off between Mengea and Mengellidae and Corioxenidae being fully resolved with the addition of Eocenoxenos to the Dundoxenos - Triazocera - Proceroxenos - Uniclavus clade. Monophyly of Mengenillidae (clade 5) is recovered, but, as already mentioned in Pohl and Beutel (2005, p. 367) and in Bravo et al. (2009: 620), it is not very well supported (Figure 6).
Stylopidae too are poorly supported in our preferred solution, as are Halictophagidae, Elenchidae and Myrmecolacidae. Nevertheless, Stylopidia (node 7) and Stylopiformia (node 17) are resolved exactly as in Pohl and Beutel (2005), although the relationships within Stylopiformia seem far from being settled, as McMahon et al. (2011) recently obtained yet another phylogeny, based on molecular evidence. Well supported nodes in the implied weighting consensus tree are node 1 (Strepsiptera), node 2, node 3, node 6 (Mengenilla), node 7 (Stylopidia), and node 8 (Corioxenidae). Within Corioxenidae, subfamily Corioxeninae as defined by Kathirithamby on the basis of four-segemented tarsi and the absence of claws (1989: 74) is paraphyletic in our cladogram and cannot be considered a natural taxon. A similar result was obtained by Pohl and Beutel (2005) and McMahon et al. (2011). On the other hand, Blissoxeninae, defined here as consisting of the genera Blissoxenos and Floridoxenos, is rather well supported on the preferred tree, confirming the findings of Pohl and Beutel (2005: 368).
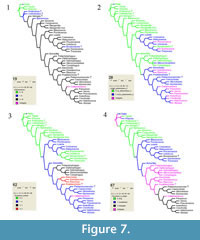 Eocenoxenos gen. n. fits well in Stylopidia (Figure 6, clade 7) by the presence of a membranous coronal suture (9:2), a wide mouthfield sclerite (52:1), and the absence of claws (63:3). Eocenoxenos also clearly belongs in Corioxenidae, sharing many autapomorphies of the family (Figure 6), as detailed in the description above. It is the sister genus of node 16, encompassing Dundoxenos Carvalho, 1956 and Triozocera Pierce, 1909, the Triozocerinae of Kathirithamby (1989: 71) and shares with them a palpus maxillaris inserted proximad of the apical stipital margin (45:1). Eocenoxenos also shares a straight aedeagus (80:0) with Triozocerinae, Uniclavinae and the genera Corioxenos and Malayaxenos within Corioxenidae. Eocenoxenos also differs from the hypothetical common ancestor of clade 15 (Triozocerinae and Eocenoxenos) by the presence of eight antennomeres (19:1) instead of less than eight (Figure 7.1), antennomere 5 as long as antennomere 3 (28:0) instead of twice as long (Figure 7.2), and a triangular (67:2) instead of isodiametric tarsomere 4 (Figure 7.4). A striking feature of Eocenoxenos is the combination of derived characteristics typical for Corioxenidae, as summarised in the description, with a character only known from Strepsiptera holding a basal position in the phylogeny: the presence of eight antennomeres (19:1), present in the fossil genera Protoxenos and Cretostylops, and regained independently in Bahiaxenos (Figure 7.1). The elaborate, extensively flabellated antennae, as well as the large compound eyes of Eocenoxenos can be considered an adaptation associated with the necessity to find females in a very short time span while being unable to feed, under environmental conditions that we are unable to reconstruct (Beutel and Pohl, 2005, p. 17).
Eocenoxenos gen. n. fits well in Stylopidia (Figure 6, clade 7) by the presence of a membranous coronal suture (9:2), a wide mouthfield sclerite (52:1), and the absence of claws (63:3). Eocenoxenos also clearly belongs in Corioxenidae, sharing many autapomorphies of the family (Figure 6), as detailed in the description above. It is the sister genus of node 16, encompassing Dundoxenos Carvalho, 1956 and Triozocera Pierce, 1909, the Triozocerinae of Kathirithamby (1989: 71) and shares with them a palpus maxillaris inserted proximad of the apical stipital margin (45:1). Eocenoxenos also shares a straight aedeagus (80:0) with Triozocerinae, Uniclavinae and the genera Corioxenos and Malayaxenos within Corioxenidae. Eocenoxenos also differs from the hypothetical common ancestor of clade 15 (Triozocerinae and Eocenoxenos) by the presence of eight antennomeres (19:1) instead of less than eight (Figure 7.1), antennomere 5 as long as antennomere 3 (28:0) instead of twice as long (Figure 7.2), and a triangular (67:2) instead of isodiametric tarsomere 4 (Figure 7.4). A striking feature of Eocenoxenos is the combination of derived characteristics typical for Corioxenidae, as summarised in the description, with a character only known from Strepsiptera holding a basal position in the phylogeny: the presence of eight antennomeres (19:1), present in the fossil genera Protoxenos and Cretostylops, and regained independently in Bahiaxenos (Figure 7.1). The elaborate, extensively flabellated antennae, as well as the large compound eyes of Eocenoxenos can be considered an adaptation associated with the necessity to find females in a very short time span while being unable to feed, under environmental conditions that we are unable to reconstruct (Beutel and Pohl, 2005, p. 17).
Eocenoxenos will most probably have parasitized on Heteroptera, as is typical for Corioxenidae. Possibly, the hosts belonged to Cydnidae (Kathirithamby, 1989: 71), a family which has been described from the Tertiary (Schaefer and Crepet, 1986; Thomas, 1988) and even from the Secondary era (Yao et al., 2007). As most other Stylopidia, Eocenoxenos lacks tarsal claws, although they are still present in its close relatives in clades 14 and 16, Uniclavinae and Triozocerinae. However, the claws in clades 14 and 16 are reduced and differ significantly from the claws in more basal Strepsiptera. If these claws would have been regained in the course of evolution, instead of being plesiomorphic, this aberrant morphology would be in accordance with Dollo's principle (Dollo, 1893).
ACKNOWLEDGEMENTS
Micro-CT scan expenses were partially supported with a grant of the KAVE (Royal Entomological Society of Antwerp), partially by the Department of Subatomic and Radiation Physics, Gent University that provided the beam time, the reconstructions, and the experimental file building for virtual and physical 3D viewing.
Thanks are due to M. Veta (Lithuania), for the cooperation in the search for new amber inclusions, to the referees for remarks and comments. H. Pohl is thanked for many detailed and interesting referee's comments which helped to significantly improve the manuscript.
REFERENCES
Agnarsson, I. and Miller, J.A. 2008. Is ACCTRAN better than DELTRAN? Cladistics 24:1032-1038.
Beutel, R.G. and Pohl, H. 2005. Head structures of males of Strepsiptera (Hexapoda) with emphasis on basal splitting events within the order. Journal of Morphology, 267:536-554.
Bosselaers J. and Jocqué R. 2000. Hortipes, a huge genus of tiny African spiders (Araneae, Liocranidae). Bulletin of the American Museum of Natural History, 256:1-108.
Bosselaers, J. and R. Jocqué. 2002. Studies in Corinnidae: cladistic analysis of 38 corinnid and liocranid genera, and transfer of Phrurolithinae. Zoologica Scripta, 31:241-270.
Bravo, F., Pohl, H., Silva-Neto, A., and Beutel, R.G. 2009. Bahiaxenidae, a "living fossil" and a new family of Strepsiptera (Hexapoda) discovered in Brazil. Cladistics, 25:614-623.
Bremer, B., Jansen, R.K., Oxelman, B., Backlund, M., Lantz, H., and Kim, K-J. 1999. More characters or more taxa for a robust phylogeny - case study from the coffee family (Rubiaceae). Systematic Biology, 48:413-435.
Bremer, K. 1988. The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. Evolution, 42:795–803.
Bremer, K. 1994. Branch support and tree stability. Cladistics, 10:295-304.
Bugnion, E., Devine, S., and Rosenblum, M. 2000. Virtual machine monitors for scalable multiprocessors. US patent 6075938.
Carvalho, L. 1956. Primeira contribuição para o Estudo dos Estresípteros angolensis (Insecta, Strespiptera). Publicaçõs Culturais da Companhia de Diamantes de Angola 29:11-54 [28].
Coddington, J.A. and Scharff, N. 1994. Problems with zero-length branches. Cladistics, 10:415-423.
Dollo, L. 1893. Les lois de l' évolution. Bulletin de la Société Belge de Géologie, de Paléontologie et d' Hydrologie, 7:164-166.
Dubois, M.B. and LaPolla, J.S. 1999. A preliminary review of Colombian ants (Hymenoptera: Formicidae) preserved in copal. Entomological News, 110:162-172.
Goloboff, P.A 1993. Estimating character weights during tree search. Cladistics, 9:83-91.
Goloboff, P.A. 1999. Analyzing large data sets in reasonable times: solutions for composite optima. Cladistics, 15:415-428.
Goloboff, P.A., Farris, J.S., and Nixon, K.C. 2003. TNT, Tree Analysis using New Technology. Version 1.1, sponsored by the Willy Hennig Society. Available from: http://www.cladistics.com (2008-12-30).
Goloboff, P.A., Farris, J.S., and Nixon, K.C. 2008b. TNT, a free program for phylogenetic analysis. Cladistics, 24:774-786.
Goloboff, P.A., Carpenter, J.M., Arias, J.S., and Miranda Esquivel, D.R. 2008a. Weighting against homoplasy improves phylogenetic analysis of morphological data sets. Cladistics, 24:758-773.
Grote, A.R. 1886. Changes Triaena Menge preoccupied, to Menge n.nov. Canadian Entomologist 18:100.
Hawkins, J.A., Hughes, C.E., and Scotland, R.W. 1997. Primary homology assessment, characters and character states. Cladistics, 13:275-283.
Henderickx, H.A.,Tafforeau, P., and Soriano C. 2012. Phase-contrast microtomography reveals the morphology of a partially visible new Pseudogarypus in Baltic amber (Pseudoscorpiones: Pseudogarypidae). Palaeontologia Electronica, 15:2,17A): 1-11. (http://palaeo-electronica.org/content/2012-issue-2-articles/252-pseudogarypus-synchrotron)
Henderickx, H., Cnudde, V., Masschaele, B., Dierick, M., Vlassebroeck, J., and Van Hoorebeke, L. 2006. Description of a new fossil Pseudogarypus (Pseudoscorpiones: Pseudogarypidae) with the use of X-ray micro-CT to penetrate opaque amber. Zootaxa, 1305:41-50.
Hofeneder K. 1910 - Stichotrema n.g. Dalla-Torreanum n.sp. Eine in einer Orthoptere lebende Strepsiptere. Zoologischer Anzeiger 36:47-49.
Hovenkamp, P. 1999. Unambiguous data or unambiguous results? Cladistics, 15:99-102.
Hovenkamp, P. 2009. Support and stability. Cladistics, 25:107-108.
Hünefeld, F., Pohl, H., Wipfler, B., Beckmann, F., and Beutel, R. 2011. The male postabdomen and genital apparatus of Mengea tertiaria, a strepsipteran amber fossil (Insecta). Journal of Zoological Systematics and Evolutionary Research, 49:298-308.
Kathirithamby, J. 1989. Review of the order Strepsiptera. Systematic Entomology, 14: 41-92.
Kathirithamby, J. and Henderickx, H., 2008. First record of the Strepsiptera genus Caenocholax in Baltic amber, with the description of a new species. Phegea, 36:149-156.
Kim, J. 1998. Large-scale phylogenies and measuring the performance of phylogenetic estimators. Systematic Biology, 47:43-60.
Kinzelbach, R. 1970. Loana Canadensis n. gen. n. sp. und die Untergliederung der Callipharixenidae (Insecta: Strepsiptera). Senckenbergiana biol. 51 (1/2):99-107.
Kinzelbach, R. 1979. Das erste neotropische Fossil der Fächerflügler (Stuttgarter Bernstainsammlung: Insecta, Strepsiptera). Stuttgarter Beiträge zur Naturkunde, serie B, Geologie und Paläeontologie 52:1-14.
Kulicka, R. 2001. New genera and species of Strepsiptera from the Baltic amber. Prace Museum Ziemi 46:3–16.
Latreille, P.A. 1802. Families naturelles et genres. Histoire naturelle, générale et particulière des crustacés et des insectes : ouvrage faisant suite aux oeuvres de Leclerc de Buffon, et partie du cours complet d'histoire naturelle rédigé, F. Dufart, Paris.
Leconte, J.L. 1874. On the Cupesidae of North America. Transactions of the American Entomological Society 5:87-88.
Linnaeus, C. 1758. Systema Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, cum Characteribus, Differentiis, Synonymis. Stockholm, Sweden.
Maddison, W.P., Donoghue, M.J., and Maddison, D.R. 1984. Outgroup analysis and parsimony. Systematic Zoology, 33:83-103.
Masschaele, B.C., Cnudde, V., Dierick, M., Jacobs, P., Van Hoorebeke, L., and Vlassenbroeck, J. 2007. UGCT. New X-ray Radiography and Tomography Facility. Nuclear Instruments and Methods in Physics Research A, 580:266-269
McMahon, D.P., Hayward, A., and Kathirithamby, J. 2011. The first molecular phylogeny of Strepsiptera (Insecta) reveals an early burst of molecular evolution correlated with the transition to endoparasitism. PLoS ONE 6:e21206.
Miyamoto, S.T. and Kifune, T. 1984. Descriptions of a new genus and two new species of Strepsiptera parasitic on Japanese Heteroptera (Strepsiptera, Corioxenidae). Kontyû, 52:137-149.
Nixon, K.C. 2002. Winclada version 1.00.08. Computer programme and documentation. Available from: http://www.cladistics.com (last accessed 30 December 2008).
Nixon, K.C. and Carpenter, J.M. 1996. On consensus, collapsibility, and clade concordance. Cladistics, 12:305-321.
Pierce, W. D. 1909. A monographic revision of the twisted winged insects comprising the order Strepsiptera Kirby. Bulletin of the United States National Museum 66:1-232.
Platnick, N.I. 2012. Less on homology. Cladistics Article first published online: 13 SEP 2012 | DOI: 10.1111/j.1096-0031.2012.00432.x
Poe, S. and Swofford, D.L. 1999. Taxon sampling revisited. Nature, 398:299-300.
Pohl, H., Beutel, R.G. 2004. Fine structure of adhesive devices of Strepsiptera (Insecta). Arthropod Structure and Development, 33:31-43.
Pohl, H. and Beutel, R.G. 2005. The phylogeny of Strepsiptera (Hexapoda). Cladistics, 21:328-374.
Pohl, H., Beutel, R.G., and Kinzelbach, R. 2005. Protoxenidae fam. nov. (Insecta, Strepsiptera) from Baltic amber - a "missing link" in strepsipteran phylogeny. Zoologica Scripta, 34:57-69.
Pohl, H., Niehuis, O., Gloyna, K., Misof, B., and Beutel, R. 2012. A new species of Mengenilla (Insecta, Strepsiptera) from Tunisia. Zookeys, 198:79-101.
Sanderson, M.J. and Donoghue, M.J. 1989. Patterns of variation in levels of homoplasy. Evolution, 42:1781–1795.
Schaefer, C.W. and Crepet, W.L. 1986. A new burrower bug (Heteroptera: Cydnidae) from the Paleocene/Eocene of Tennessee. Journal of the New York Entomological Society, 94:296-300.
Sorenson, M.D. and Franzosa, E.A. 2007. TreeRot.v3. Computer programme and documentation. Available from: http://people.bu.edu/msoren/TreeRot.html (last accessed 26 November 2012).
Swofford, D.L. (2002) PAUP*Phylogenetic Analysis under Parsimony (*and other methods) version 4.0 beta 10. Sinauer Associates, Sunderland, MA.
Thomas, D.B. 1988. Fossil Cydnidae (Heteroptera) from the Oligo-Miocene amber of Chiapas, Mexico. Journal of the New York Entomological Society, 96:26-29.
Vlassenbroeck, J., Dierick, M., Masschaele, B., Cnudde, V., Van Hoorebeke, L., and Jacobs, P. 2007. Software tools for quantification of X-ray microtomography at the UGCT. Nuclear Instruments and Methods in Physics Research, A 580:442-445.
Watrous, L.E. and Wheeler, Q.D. 1981. The out-group comparison method of character analysis. Systematic Zoology, 30:1-11.
Wilkinson, M. 1995. Arbitrary resolutions, missing entries, and the problem of zero-length branches in parsimony analysis. Systematic Biology, 44:108-111.
Yao, Y., Cai, W., and Ren, D. 2007. The first fossil Cydnidae (Hemiptera: Pentatomoidea) from the late Mesozoic of China. Zootaxa, 1388:59-68.

