 Drishti and Amira – different visualizations exemplified by the early Cambrian Chengjiang arthropod Leanchoilia illecebrosa from China
Drishti and Amira – different visualizations exemplified by the early Cambrian Chengjiang arthropod Leanchoilia illecebrosa from China
Article number: 28.1.a5
https://doi.org/10.26879/1358
Copyright Palaeontological Association, January 2025
Author biographies
Plain-language and multi-lingual abstracts
PDF version
Submission: 13 November 2023. Acceptance: 18 December 2024.
ABSTRACT
Virtual paleontology plays a pivotal role in elucidating the morphology of ancient arthropods. Since 2015, the open-source volume rendering software Drishti, has been employed to uncover concealed morphological features preserved within fossils dating back to the early Cambrian Chengjiang biota, approximately 518 million years old and located in southwest China. Also another program, Amira, has emerged as a potent tool for such investigations. In this study, we undertake a comparative analysis of both programs to comprehensively assess their technical capabilities regarding visualization modes and associated challenges.
We selected the extensively studied megacheiran arthropod, Leanchoilia illecebrosa, as a model species and demonstrated how various rendering options, including volume rendering, surface rendering, and isosurface rendering, can be implemented in each program. Furthermore, we showcased the visual outcomes of specific operations such as slicing (utilizing clipping planes and orthoslices) as well as direct volumetric modifications (MOP carve and paint). Our findings indicate that Drishti excels in volume visualization due to its ability to produce realistic and organic graphical representations. In contrast, Amira offers rapid generation of (iso)surface models, which can serve as meshes for subsequent 3D model-based analyses. Both programs prove to be suitable for the analysis of pyritized fossils and enable the depiction of diverse morphological visualizations. However, it is important to note that each program possesses its own set of advantages and limitations. This comparative study aims to explore these characteristics and provide a valuable resource for researchers in selecting the most appropriate tool for their specific scientific purposes.
Michel Schmidt. Yunnan Key Laboratory for Palaeobiology, Yunnan University, North Cuihu Road 2, Kunming 650091, People’s Republic of China and MEC International Joint Laboratory for Palaeobiology and Palaeoenvironment, Yunnan University, 2 North Cuihu Road, Kunming 650091, People’s Republic of China and Ludwig-Maximilians-Universität München, Biocenter, Großhaderner Str. 2, 82152 Planegg-Martiensried, Germany and MEC International Joint Laboratory for Palaeobiology and Palaeoenvironment, Yunnan University, 2 North Cuihu Road, Kunming 650091, People’s Republic of China. michel.m.schmidt@outlook.com
Roland R. Melzer. MEC International Joint Laboratory for Palaeobiology and Palaeoenvironment, Yunnan University, 2 North Cuihu Road, Kunming 650091, People’s Republic of China and Bavarian State Collection of Zoology, Bavarian Natural History Collections, Münchhausenstr. 21, 81247 Munich, Germany and Ludwig-Maximilians-Universität München, Biocenter, Großhaderner Str. 2, 82152 Planegg-Martiensried, Germany and GeoBio-Center, Ludwig-Maximilians-Universität München, Luisenstr. 37, 80333 Munich, Germany. melzer@snsb.de
Huijuan Mai. Yunnan Key Laboratory for Palaeobiology, Yunnan University, North Cuihu Road 2, Kunming 650091, People’s Republic of China and MEC International Joint Laboratory for Palaeobiology and Palaeoenvironment, Yunnan University, 2 North Cuihu Road, Kunming 650091, People’s Republic of China. Huijuan.Mai@ynu.edu.cn
Xianguang Hou. Yunnan Key Laboratory for Palaeobiology, Institute of Palaeontology, Yunnan University, 650500 Kunming, Yunnan, China and MEC International Joint Laboratory for Palaeobiology and Palaeoenvironment, Yunnan University, 650500 Kunming, Yunnan, China. xghou@ynu.edu.cn
Yu Liu. Yunnan Key Laboratory for Palaeobiology, Yunnan University, North Cuihu Road 2, Kunming 650091, People’s Republic of China and MEC International Joint Laboratory for Palaeobiology and Palaeoenvironment, Yunnan University, 2 North Cuihu Road, Kunming 650091, People’s Republic of China. (Corresponding author) Yu.liu@ynu.edu.cn
Keywords: arthropod; virtual palaeontology; Leanchoilia; comparison; Cambrian; morphology
Final citation: Schmidt, Michel, Melzer, Roland R., Mai, Huijuan, Hou, Xianguang, and Liu, Yu. 2025. Drishti and Amira – different visualizations exemplified by the early Cambrian Chengjiang arthropod Leanchoilia illecebrosa from China. Palaeontologia Electronica, 28(1):a5.
https://doi.org/10.26879/1358
palaeo-electronica.org/content/2025/5401-drishti-and-amira-visualizations
Copyright: January 2025 Palaeontological Association.
This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
creativecommons.org/licenses/by/4.0
INTRODUCTION
Virtual paleontology involves the study of fossils through interactive digital visualizations and employs non-invasive, non-destructive methods to reveal the internal anatomical structures of fossils (Garwood et al., 2010; Cunningham et al., 2014; Sutton et al., 2014; Yin and Lu, 2019). Computed tomography (CT), one of the techniques, has proven to be an essential tool for examining morphology and hidden structures, making it one of the most used techniques in virtual paleontology for analyzing suitable fossils (Sutton et al., 2014).
In particular, the application of micro-computed tomography (μ CT), a specialized form of CT designed for smaller objects, has significantly advanced the understanding of the morphology of numerous invertebrate fossils over the past decade (Liu et al., 2015, 2016, 2020a, b, 2021; Chen et al., 2019; Zhai et al., 2019a, b, c, 2022; Ou et al., 2020; Jin et al., 2021; Schmidt et al., 2020, 2021a, 2022a, 2024; Zhang, M. et al., 2022; Zhang, X. et al., 2022; O’Flynn et al., 2024). However, visualizing 3D volumetric data obtained from μ CT scans poses certain challenges, leading to the development of various protocols for this purpose (e.g., Semple et al., 2019; Buser et al., 2020), which have influenced the direction of virtual paleontology (Sutton et al., 2014; Keklikoglou, 2019). A wide range of open-source (e.g., SPIERS, IrfanView, ParaView, ImageJ, 3DSlicer, Blender, MeshLab) and commercial (e.g., VG Studio Max, Mimics, Imaris, VoluMedics) software applications for 3D visualization are available. Most of these programs are well-equipped for image segmentation and rendering both volume and surface models. The selection of specific software often depends on factors such as the availability of programs at different research institutions, the budget aspect, and first of all the type of fossil preservation.
The renowned Chengjiang Lagerstätte, a remarkable fossil site located in Southwest China and dating back 518 million years, is recognized for its exceptional fossil preservation (Gaines et al., 2012; Hou et al., 2017). This extraordinary preservation allows for detailed X-ray computed 3D studies in the realm of virtual paleontology (Liu et al., 2015). Recent research has uncovered intricate 3D structures within various arthropods from Chengjiang, which are vital for understanding their limb morphology and evolutionary history (Zhao et al., 2017; Chen et al., 2019; Zhai et al., 2019a, b, 2022; Liu et al., 2021; Schmidt et al., 2020, 2021a, 2022a, 2024; Zhang, M. et al., 2022; Zhang, X. et al., 2022), as well as their ontogeny and ecological roles (Liu et al., 2016, 2020a; Zhai et al., 2019c; O’Flynn et al., 2024). Most of these investigations have employed an open-source software called Drishti, which consists of three components (Drishti Import, Drishti Paint, and the Drishti renderer) and provides a variety of volume rendering techniques and tools, enhancing the realistic and organic appearance of scanned and volume-rendered specimens (Limaye, 2012; Hu et al., 2020, 2024).
Amira (Thermo Fisher Scientific, 2018) is another useful 3D visualization software for processing μ CT data (Stalling et al., 2005; Ruthensteiner, 2008). It has been utilized to uncover new morphological characteristics in Cambrian arthropods (Cheng et al., 2011), particularly concerning Chengjiang arthropods (Liu et al., 2015; Schmidt et al., 2020, 2021a). However, it is important to note that this software is commercial, unlike Drishti. The financial aspect should be considered when deciding which program to use for μ CT data analysis.
This program has nonetheless established a strong reputation in biological research (Kudryashev et al., 2011, 2012; Meisslitzer-Ruppitsch et al., 2011) and has played a crucial role in reconstructing the morphology of both living (Frase and Richter, 2013; Jaszkowiak et al., 2015; Keiler et al., 2015) and extinct species (Gunz et al., 2009; Lukeneder et al., 2014), enabling the visualization of both internal and external anatomical features. Drishti and Amira are software applications specifically designed for the scientific visualization of volumetric data. They transform two-dimensional image data, such as TIFF files, into three-dimensional volumes, which can then be manipulated through cropping, slicing, painting, and converting into surface models. It is important to note that each program has its own unique advantages and limitations. Our main goal is to provide an overview of these differences, serving as a valuable resource to help researchers choose the most suitable tool for their specific scientific needs.
For our comparative approach on visualization, we have selected the extensively studied megacheiran arthropod, Leanchoilia illecebrosa (Liu et al., 2007, 2014, 2016, 2020b, 2021), as our model species. We employ this model to juxtapose the various procedures involved in rendering, aiming to delineate discrepancies in the visualization outcomes achieved through the utilization of Drishti and Amira, and we elucidate disparities in the graphical outputs. Additionally, we outline the general processing steps necessary for creating 3D models of fossils from μ CT data using Drishti, as this software is more commonly used compared to Amira in the μ CT-based analysis of the ventral morphology of arthropods from the Chengjiang biota.
MATERIALS AND METHODS
Drishti (Limaye, 2012) and Amira (Thermo Fisher Scientific, 2018) renderings are demonstrated using two specimens of Leanchoilia illecebrosa from the Chengjiang biota. These specimens were obtained from the Yu´ anshan Member, Chiungchussu Formation, Eoredlichia-Wutingaspis trilobite biozone, Cambrian Series 2, Stage 3, Yunnan Province, China, and are housed at the Yunnan Key Laboratory for Palaeobiology in Kunming, China. While Specimen YKLP 11422-arthro has been previously documented in Liu et al. (2020b), the other specimen, YKLP 11439-arthro, is presented for the first time in this study. The μ CT scans of these specimens were conducted at the Yunnan Key Laboratory for Palaeobiology, Kunming, utilizing the Xradia 520 Versa system (Carl Zeiss X-ray Microscopy, Pleasanton, USA). The scanning parameters included a beam strength of 40 kV/3 w, a LE3 filter, and a resolution of 458 µm. Volumetric data were stored in TIFF stacks.
We employed Drishti version 2.7 (https://github.com/nci/drishti/releases) and Amira version 6.3 (https://www.thermofisher.com/de/de/home/electron-microscopy/products/software-em-3d-vis/amira-software.html) for our data processing. In Drishti, the TIFF stacks were imported into Drishti Import to produce a. pvl.nc file, which was subsequently loaded into Drishti renderer. The volumetric data were processed in high-resolution mode, with Transfer Functions optimized for the best visualization properties. Segmentation of individual appendages was carried out after loading the.pvl.nc file into Drishti Paint, employing various tools. The exported files were reloaded into Drishti renderer for visualization purposes.
In Amira, the TIFF stacks were directly imported into the program’s main interface. For specific tasks, the respective fields were linked to the TIFF stack. Amira version 6 provides two distinct modules for volume visualization: the “Volren” and the “Volume Rendering” modules, both of which facilitate volume visualization. In this study, we opted for the “Volren” module for volume rendering.
To clarify the key technical terms in the context of scientific data visualization, we provide the following definitions (Thermo Fisher Scientific, 2018):
Volume rendering (“Volren”): This term encompasses a range of techniques applied to 3D data sets (scalar fields) to project them as 2D projections.
Isosurface rendering: This method offers a rapid means of reconstructing polygonal surface models of the entire object, enabling the analysis of arbitrary scalar fields sampled on distinct grids.
Surface rendering: This process involves the manual or (semi-)automatic generation of mesh models for specific regions through segmentation, using various tools (e.g., brush, lasso, magic wand, threshold, etc.)
Segmentation: This refers to the assignment of labels to image voxels, aiding in the identification and separation of objects within a three-dimensional image.
RESULTS AND DISCUSSION
Processing Features: a Comparison
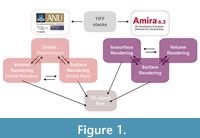 In our examination of both Drishti and Amira, we first highlight disparities in the processing procedures (Figure 1) before delving into a detailed comparison of their respective visualization modes and mesh reconstruction capabilities.
In our examination of both Drishti and Amira, we first highlight disparities in the processing procedures (Figure 1) before delving into a detailed comparison of their respective visualization modes and mesh reconstruction capabilities.
Processing in Drishti. In the case of Drishti, volumetric data, which may include TIFF stacks or any of the 13 supported volumetric data formats, is initially imported into the dedicated Drishti Import module for conversion into.pvl.nc files (processed volume files). In contrast, Amira permits the direct import of volumetric data, either through a simple drag-and-drop process or by importing the data directly into the program. In both instances, specifying the voxel size proves advantageous, although this parameter can also be adjusted subsequently, either in the crop editor (Amira) or in the settings (Drishti).
Following this initial step, Drishti-generated.pvl.nc files are further processed within the Drishti renderer itself. This environment offers a range of options, including morphological operations (MOP), measurement tools, and the ability to adjust clipping planes, among others. Notably, these operations primarily pertain to work in volume rendering mode. The resultant surface triangle-mesh formats can subsequently be exported from the Drishti renderer in various file formats (e.g.,.ply or.stl files).
Conversely, in the Drishti Import,.pvl.nc files generated earlier can be transferred to another distinct component within the Drishti software suite, namely Drishti Paint. This specialized program focuses on surface rendering and allows for the segmentation of object regions in both 2D (xy, xz, yz planes) and 3D (xyz). The segmented regions can be exported as either.pvl.nc files (for re-import into the Drishti renderer) or as mesh formats such as .ply or .stl files, which can be further processed in software applications like MeshLab, Blender, Maya, or various other programs.
Processing in Amira. Amira, functioning as a standalone program, provides analogous options, albeit within a unified environment. Upon importing volumetric data, users can select the project tab and assign attributes to the data label field, such as an isosurface label field. By adjusting threshold values and considering options like compactification and downsampling to reduce data size, one can swiftly visualize the volumetric model as an automatically surface-rendered model. An alternative approach for surface model visualization in Amira involves the segmentation of regions of interest, a task performed in the segmentation editor. These labels can subsequently be transformed into surfaces and extracted as 3D objects (meshes) in various file formats.
Furthermore, Amira extends the capability for pure volume rendering, which can be accomplished in the project tab after assigning the “Volren” label to the volumetric data label. Additional functionalities, such as orthoslices, measurements, and various shaders, can be applied to the volume rendering process.
Visualization Modes: a Comparison
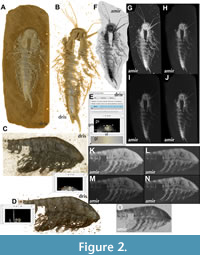 Volumetric visualizations. Volume rendering stands as a central feature within the Drishti renderer. Upon importing the .pvl.nc files, derived from the initial volumetric data processed in Drishti Import, a critical task is to determine the optimal transfer function, enabling a clear and accurate depiction of the fossil (Figure 2A, B). The effectiveness of this process hinges upon factors such as the quality of the fossil specimen and the resolution of the scanned object (Figure 2C, D). Thus, the meticulous adjustment of transfer functions assumes paramount importance when working with Drishti, underscoring the significance of understanding the intricacies of the transfer function editor (Figure 2E).
Volumetric visualizations. Volume rendering stands as a central feature within the Drishti renderer. Upon importing the .pvl.nc files, derived from the initial volumetric data processed in Drishti Import, a critical task is to determine the optimal transfer function, enabling a clear and accurate depiction of the fossil (Figure 2A, B). The effectiveness of this process hinges upon factors such as the quality of the fossil specimen and the resolution of the scanned object (Figure 2C, D). Thus, the meticulous adjustment of transfer functions assumes paramount importance when working with Drishti, underscoring the significance of understanding the intricacies of the transfer function editor (Figure 2E).
In the plotting interface, six default points are provided, collectively forming the “spine.” This region’s size can be expanded or reduced by individually dragging and dropping each point or by manipulating all points simultaneously along the horizontal axis (representing voxel intensity) or the vertical axis (representing gradient magnitude). The plotting interface is available in both 1D and 2D views, and all settings can be preserved, much like in the gradient interface where color schemes can be switched. These settings can be saved by pressing ‘S’ while hovering over the interface and later reactivated by pressing the space bar to access the transfer function library. This procedural step holds foundational importance, as it obviates the need for iterative modification of transfer functions to recreate a specific visualization aspect.
Volume rendering in Amira employs a distinct approach. After importing the volumetric data into the main window, the assignment of the “Volren” label field to the data field is required, which in turn generates a volume file. This volume file can be visualized in four different modes, based on two major approaches, namely Maximum Intensity Projection (MIP) and Texture-Based Volume Rendering (VRT).
The MIP volume rendering mode presents the brightest data value along each ray of sight, in contrast to displaying the outcome of the emission absorption model. It can be visualized in either an inverted (Figure 2F, O) or a normal (Figure 2G, K) orientation. On the other hand, the VRT mode, as a texture-based volume rendering, offers various shading options, including diffuse or specular, alongside the standard one (Figure 2H, L). The diffuse VRT mode (Figure 2I, M) employs a diffuse light source, while the specular VRT option (Figure 2J, N) simulates a specular visualization of the specimen. The latter closely resembles the presets of Drishti’s volume rendering, resulting in a more realistic appearance. However, as in Drishti, the efficacy of these rendering modes is contingent upon the quality of the fossil. In our example, despite specimen YKLP 11439-arthro having a resolution of 458 µm, it fails to reveal extensive morphological information in comparison to specimen YKLP 11422-arthro. Altering the transfer function, i.e., modifying the width and height of the spine, for specimen YKLP 11439-arthro still does not yield a similar visualization to that of specimen YKLP 11422-arthro. When encountering such challenges during volume rendering, irrespective of the program being used, it may be necessary to consider rescanning the fossil with different settings or opting for a different specimen altogether. After all, despite having sufficient experience, it is sometimes not immediately apparent from the outside whether a fossil has enough morphological structures preserved inside to make qualitative statements about the nature and composition of those structures.
In a direct comparison of volume rendering visualization methods between Drishti and Amira, the MIP mode in Amira stands out as the swiftest means for an initial comprehensive visualization of fossils. By highlighting the brightest data value along each ray of sight, it occasionally allows for an almost transparent view of the fossil when observed dorsally, unveiling the underlying anatomical structures. In Drishti, the initial challenge often lies in the precise determination of suitable transfer functions, particularly when dealing with pyritized fossils entwined with surrounding rock matrices, a more intricate task than rendering fossilized bones devoid of such matrix. The superior performance between MIP and VRT modes is also contingent upon the quality of the fossil. In our illustrative cases, specimen YKLP 11422-arthro manifests the most discernible features in MIP mode (Figure 2F, G), while specimen YKLP 11439-arthro delivers detailed outcomes in both MIP (Figure 2K, O) and VRT specular (Figure 2N). In the case of the latter specimen, we would recommend employing Amira over Drishti, especially when precision in rendering morphological details is essential, as the visualization process in Drishti often necessitates a significant degree of “trial and error” to achieve the optimal transfer function settings for clear depiction of anatomical features (Figure 2C, D).
Cross-sections: clipping planes and orthoslices. Drishti and Amira both offer tools for conducting cross-sectional examinations of volumetric renderings, utilizing clipping planes in Drishti and orthoslices in Amira (also known as the “Slice Module” or “Oblique Slice” in earlier versions). These tools allow users to explore fossils by projecting 2D TIFF slices onto 3D datasets, enabling the isolation of specific areas for detailed analysis.
While both tools share similarities, such as the ability to slice through datasets and enhance visualization, they differ in functionality. Drishti's clipping planes are interactive and can be adjusted to create various cuts, allowing for the simultaneous use of multiple planes. In contrast, Amira's orthoslices are more static, aligned along the X, Y, and Z axes, and primarily limited to these three main orientations. Additionally, the user interfaces and control mechanisms for these features reflect the distinct design choices of their developers.
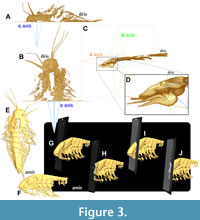 In Drishti, users can activate clipping planes by pressing “c” while hovering over the volume. The axes are represented as the a-axis (orange), b-axis (green), and c-axis (blue) (Figure 3A, B). These planes segment the fossil into left and right, anterior and posterior, as well as dorsal and ventral sections. The default clipping plane typically corresponds to the frontal plane, depending on the fossil’s orientation. By hovering over the volume, pressing the spacebar to access the command window, and entering “rotateb 180,” a sagittal section of the fossil can be obtained (Figure 3A, B). This method, illustrated with specimen YKLP 11422-arthro, can provide valuable insights into the structure and composition of the basal part of the great appendage (Figure 3C, D). The clipping plane can also be adjusted along the chosen axis, and users have the option to freely rotate around all axes to any extent. Furthermore, they can modify the “rotate” command by adding “-b” or “-c” to the term “rotate” and specifying the desired angle (for instance, the command “rotatec 60” rotates the model by 60° around the c-axis).
In Drishti, users can activate clipping planes by pressing “c” while hovering over the volume. The axes are represented as the a-axis (orange), b-axis (green), and c-axis (blue) (Figure 3A, B). These planes segment the fossil into left and right, anterior and posterior, as well as dorsal and ventral sections. The default clipping plane typically corresponds to the frontal plane, depending on the fossil’s orientation. By hovering over the volume, pressing the spacebar to access the command window, and entering “rotateb 180,” a sagittal section of the fossil can be obtained (Figure 3A, B). This method, illustrated with specimen YKLP 11422-arthro, can provide valuable insights into the structure and composition of the basal part of the great appendage (Figure 3C, D). The clipping plane can also be adjusted along the chosen axis, and users have the option to freely rotate around all axes to any extent. Furthermore, they can modify the “rotate” command by adding “-b” or “-c” to the term “rotate” and specifying the desired angle (for instance, the command “rotatec 60” rotates the model by 60° around the c-axis).
Orthoslices in Amira can be generated by assigning the orthoslice field to the volumetric data file within the project tab. These slices are represented by three axes: xy, xz, and yz. Users can observe the orthoslices within the volume rendering and utilize them alongside isosurfaces (Figure 3E-J). Isosurfaces are quickly generated polygonal surface models that allow viewers to adjust the threshold, resulting in a variable number of triangles to enhance the presentation of fossils. When the goal is to visualize basic polygonal surface models, isosurface reconstructions offer a more straightforward method compared to segmentation (as shown below). By allocating volumetric data to the model, users can select specific TIFF slices for future reference. Amira provides various graphical modes, such as rendering TIFF slices transparent (transalpha), which can either display the anterior part (Figure 3G) or exclude it (Figure 3H), allowing users to focus on internal structures. Alternatively, the TIFF slice can be shown in parallel (transbinary; Figure 3I, J). In Amira, orthoslices maintain their perpendicular and horizontal orientation, as suggested by their name. While multiple xy-orthoslices can be applied to the model, their orientation relative to the world center of coordinates remains fixed and cannot be altered. However, users can utilize alternative non-orthogonal slices by selecting a different field for assignment to the volume.
In summary, both Clipping Planes in Drishti and Orthoslices in Amira aid users in the detailed exploration of 3D datasets. However, they offer distinct functionalities and operational characteristics that cater to varying user needs and preferences in visualizing complex data.
Carving: MOP and volume edit. In both programs, it is possible to carve within the object and isolate elements. In Drishti, this feature is available in the “Morphological Operation” module through the “MOP carve” function. In Amira, this can be done using the “Volume Edit” module.
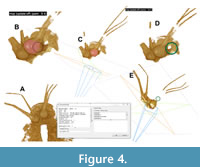 Drishti provides unique features for comprehensive volume rendering exploration, including two main capabilities: direct carving within the volumetric model to isolate and segment specific structures, and the ability to apply various colors directly onto the volumetric model to highlight areas of interest (Figure 4). Both functionalities are accessed via the morphological operation command (MOP).
Drishti provides unique features for comprehensive volume rendering exploration, including two main capabilities: direct carving within the volumetric model to isolate and segment specific structures, and the ability to apply various colors directly onto the volumetric model to highlight areas of interest (Figure 4). Both functionalities are accessed via the morphological operation command (MOP).
To initiate a MOP in Drishti, users must hover over the volumetric model and press the spacebar to open the command window. First, the viewer should input “mop update off” (Figure 4A). After that, users can start painting by using the command “MOP paint” in combination with the Shift key and left-click (Figure 4B). The colors and the radius of the painting area can be adjusted using the arrow keys, and this feature can also be integrated with clipping planes (Figure 4C). The “MOP carve” command enables users to remove specific morphological areas from the entire volume model (Figure 4D). Users can modify the settings using the arrow keys. A practical method for eliminating unwanted backscattering and non-fossil objects in the scan is to combine clipping planes with the “MOP carve” function (Figure 4E). Any actions taken in “MOP paint” and “MOP carve” can be reversed by pressing Shift and right-click.
In contrast, Amira provides comparable functionalities through its “Volume Edit” module when utilized with “Volren,” which operates similarly to Drishti'’ carving tool (MOP carve). The “Volume Edit” module in Amira version 6.3 is a robust tool designed for advanced manipulation and analysis of volumetric data. It enables users to perform tasks such as segmentation, filtering, and morphological edits to isolate features. This module supports a non-destructive workflow, allowing users to experiment with edits without permanently modifying the original dataset. The integration with the “Volren” rendering engine enhances visualization, producing high-quality representations of the edited volumes. Users can export their modifications as separate TIFF stacks, making it easy to share and conduct further analysis while minimizing background scattering for better clarity. Additionally, the module includes tools for adjusting opacity and color mapping, which facilitate more informative visualizations that emphasize specific features within the data.
Measurements. Both Drishti and Amira offer measurement capabilities, contingent upon the assigned voxel size. In Amira, the voxel size must be specified during the initial upload of volumetric data but can subsequently be adjusted. In Drishti, voxel size settings can be modified within the Drishti renderer. However, a significant divergence exists between the two programs regarding measurement capabilities. Notably, in Amira, measurements cannot be performed within the volume rendering itself. To facilitate measurements, it is imperative to have at least a generated isosurface model as a reference. Distances can be calculated by selecting regions of interest on the segmentation-based surface model or the isosurface model, utilizing the 3D ruler tab. It is crucial to configure the view to orthographic mode rather than perspective before conducting measurements in Amira. In contrast, Drishti permits measurements to be conducted directly within the volume rendering interface. To perform measurements, the viewer creates two points, presses the spacebar, and enters “path.” Subsequently, the distance in the specified unit (e.g., micron) is displayed when hovering over the line connecting the two points.
Segmentation and Surface Reconstruction: a Comparison
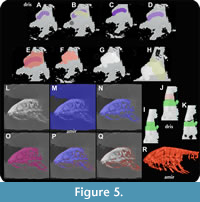 Segmentation in 2-dimensional planes (xy, xz, yz). In addition to their volume rendering capabilities, both programs offer features for segmentation and the extraction of regions of interest. Drishti utilizes a separate program called Drishti Paint for segmentation, where users can upload the .pvl.nc files generated in Drishti Import. Two-dimensional image segmentation is achieved through a process referred to as “tagging” (Figure 5A-K). Tagging involves directly painting slices of the volumetric data in all three interfaces (xy, xz, yz). A palette of 255 color tags is available, with the final one designated as the “trash tag,” used for deleting markings. Tagging is conveniently executed by hovering over the region of interest and left clicking (Figure 5A). These marked areas can also be overwritten with a different color tag (Figure 5B), and the selected color tag can be confirmed by pressing “P,” slightly darkening the color (Figure 5C). However, after confirmation, neither can the color tag be deleted using standard commands like “esc” or “delete,” nor can the region be overpainted with another color tag (Figure 5D). To erase a color-marked area, the user needs to overpaint it with tag 255, the “trash tag” (Figure 5E). After confirming the “trash tag” by pressing “P” (Figure 5F), the user can delete the entire area marked with the “trash tag” by pressing “delete” or “esc” (Figure 5G), enabling the commencement of a new painting with another color tag (Figure 5H). The “trash tag” is especially valuable when the user encounters situations involving the overlapping of two distinct structures that require different color tags (Figure 5I). In this scenario, the “trash tag” can be applied to the overlapped region by pressing Shift + left-click (Figure 5J), after which the “trash tag” can be deleted (Figure 5K).
Segmentation in 2-dimensional planes (xy, xz, yz). In addition to their volume rendering capabilities, both programs offer features for segmentation and the extraction of regions of interest. Drishti utilizes a separate program called Drishti Paint for segmentation, where users can upload the .pvl.nc files generated in Drishti Import. Two-dimensional image segmentation is achieved through a process referred to as “tagging” (Figure 5A-K). Tagging involves directly painting slices of the volumetric data in all three interfaces (xy, xz, yz). A palette of 255 color tags is available, with the final one designated as the “trash tag,” used for deleting markings. Tagging is conveniently executed by hovering over the region of interest and left clicking (Figure 5A). These marked areas can also be overwritten with a different color tag (Figure 5B), and the selected color tag can be confirmed by pressing “P,” slightly darkening the color (Figure 5C). However, after confirmation, neither can the color tag be deleted using standard commands like “esc” or “delete,” nor can the region be overpainted with another color tag (Figure 5D). To erase a color-marked area, the user needs to overpaint it with tag 255, the “trash tag” (Figure 5E). After confirming the “trash tag” by pressing “P” (Figure 5F), the user can delete the entire area marked with the “trash tag” by pressing “delete” or “esc” (Figure 5G), enabling the commencement of a new painting with another color tag (Figure 5H). The “trash tag” is especially valuable when the user encounters situations involving the overlapping of two distinct structures that require different color tags (Figure 5I). In this scenario, the “trash tag” can be applied to the overlapped region by pressing Shift + left-click (Figure 5J), after which the “trash tag” can be deleted (Figure 5K).
Segmentation in Amira follows a different approach. Users can perform segmentation using the segmentation editor and the multi-planar viewer. Amira employs “labels” and “label fields” instead of color tags, offering greater flexibility in color selection, and allowing for the deletion of label fields when a specific color is no longer desired. This flexibility is advantageous, as in Drishti, applied and confirmed color tags cannot be changed to a different color. Amira provides several tools for painting and segmenting areas of interest within the 2D data slices (xy, xz, yz planes). The “brush tool,” like color tagging in Drishti, enables users to directly paint individual voxels. The “magic wand” tool allows users to select all voxels with the same grey values within a predefined area, thereby performing a region-growing operation. The “lasso” tool permits users to draw a line around specific fields of interest and mark them by creating a closed contour curve. The “threshold tool” is the quickest and most used tool when rapid and comprehensive segmentations are required. It selects voxels through threshold segmentation by specifying minimum and maximum image intensity (Figure 5L-Q). Ultimately, the label field containing all voxel information can be used for surface reconstruction in the main window (Figure 5R). Users can create a new label field for every color or area of interest within the slices, and all label fields can be used for surface generation. This essential feature has also been incorporated into Drishti since version 2.7, enhancing the intuitiveness and speed of segmentation in Drishti Paint (for a comprehensive understanding, see Hu et al., 2020).
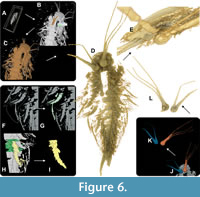 One of the most user-friendly and intuitive features in Drishti is the ability to directly paint onto the 3D volume within the volume interface using Drishti Paint (Figure 6A). In contrast, this feature is absent in Amira, where the 3D volume window is provided alongside the three slice interfaces (xy, xz, yz), but without any additional segmentation capabilities. In Drishti, users can tag the volume's surface with a simple left-click (Figure 6B) and confirm the color by pressing “F” (Figure 6C). These tagged regions, both collectively and individually, can then be extracted as .pvl.nc files and re-imported into the Drishti Renderer, resulting in a cleaned-up version of the fossil (Figure 6D). This process greatly enhances the visualization of areas that were previously challenging to examine due to background scattering (Figure 6E).
One of the most user-friendly and intuitive features in Drishti is the ability to directly paint onto the 3D volume within the volume interface using Drishti Paint (Figure 6A). In contrast, this feature is absent in Amira, where the 3D volume window is provided alongside the three slice interfaces (xy, xz, yz), but without any additional segmentation capabilities. In Drishti, users can tag the volume's surface with a simple left-click (Figure 6B) and confirm the color by pressing “F” (Figure 6C). These tagged regions, both collectively and individually, can then be extracted as .pvl.nc files and re-imported into the Drishti Renderer, resulting in a cleaned-up version of the fossil (Figure 6D). This process greatly enhances the visualization of areas that were previously challenging to examine due to background scattering (Figure 6E).
The entire 3D segmentation process in Drishti Paint is reminiscent of the use of the threshold and magic wand tools in Amira to select interconnected regions. However, as previously mentioned, these tools are limited to 2D segmentation in Amira. In Drishti, similar outcomes can be achieved through the “F” confirmation of color tags applied to a region, which segments everything connected to it. This can also be extracted as a new .pvl.nc file. Another tool, introduced in Drishti 2.7 (Hu et al., 2020), is the gradient threshold function. This thresholding tool encompasses three different gradient types and selects everything with the same voxel parameters.
In the first case, it’s essential to ensure that certain areas with no connection to the tagged and confirmed region are also tagged (for example, a biramous appendage). In the second case, one must ensure that only regions of interest of the fossil itself are selected through thresholding, excluding background scattering with similar voxel parameters.
Furthermore, Drishti Paint allows users to segment specific regions of interest by drawing lines using the sketch pad mode (with Shift + left-click to draw and “esc” to delete the line). This feature is particularly valuable when, as shown in Figure 6F, G, only specific appendicular details need to be highlighted and tagged. Regularly painting these regions with a left-click would be more time-consuming. In Amira, a similar function to the sketch pad is the previously mentioned lasso tool. This tool is a quick way to select regions of interest by drawing a line around them, but it is not as straightforward when applied to the 3D model compared to other segmentation options. Nevertheless, in some cases, such as the need to quickly cut out regions in 3D (e.g., segmented objects not belonging to the fossil), the lasso tool is more efficient than deselecting areas in 2D.
Moreover, Drishti Paint allows for segmentation of the 3D volumetric file using different color tags to separate structures from one another (Figure 6H-K). Ultimately, these tagged regions can be extracted and re-imported into the Drishti renderer, revealing hidden morphological details of previously obscured body parts (Figure 6L).
Both programs are capable of calculating and extracting surfaces from the given volume. While this all occurs within a single program in Amira, it is distributed across two programs in Drishti: the Drishti renderer and Drishti Paint.
In Drishti, the primary emphasis lies on working with volumes in the Drishti renderer program. The other program, Drishti Paint, allows users to segment data, as discussed earlier, to eliminate background noise that does not pertain to the fossil Subsequently, the segmented regions can be exported as pvl.nc files and re-imported into the Drishti renderer. In both the Drishti renderer and Drishti Paint, users have the capability to generate meshes as surfaces and export them as .ply or .stl files. The most notable advantage of mesh generation in Drishti, compared to this process in Amira, is as follows: in cases where a fossil lacks preserved original structures, while, for instance, only a cavity remains without any organic or inorganic material, the ability to view and represent these fossils is restricted to volume rendering mode. In contrast, Drishti offers the unique capability to directly transform pure volume information into surface information. This stands in contrast to Amira, where this transformation is contingent upon segmentation.
Amira offers two primary methods for surface modeling. Firstly, users can create isosurface models (see Figure 3E-J), which can be a relatively rapid process depending on the data volume. This results in the extraction of a surface from the volume, with the ability to adjust mesh density and resolution. Amira also provides the capability to measure morphological structures on these models, a feature not available when exclusively using pure volume rendering. The alternative, although somewhat time-consuming, method for obtaining a complete or partial surface model of a specific object involves image segmentation, as in the aforementioned chapter. In the segmentation editor, the viewer can segment slice by slice or across multiple slices using the interpolation function. Isosurface modeling offers time savings, creating surfaces with a consistent thickness, ultimately leading to a more accurate and unbiased surface reconstruction when compared to manually adding layers during surface modeling based on image segmentation.
Surface rendering in Amira provides a substantial advantage when working with μ CT data from Chengjiang fossils. Extracted surfaces can be seamlessly integrated into a variety of software applications, including Blender ( www.blender.org) and Autodesk Maya ( www.autodesk.com/products/maya/over-view), for advanced post-processing tasks such as modeling (Garwood & Dunlop, 2014) and kinematic studies (Schmidt et al., 2021b, 2022b).
CONCLUSIONS
In recent years, numerous Chengjiang euarthropods have undergone visualization using Drishti, shedding light on various aspects of their anatomy. These aspects include their precise ventral features (Chen et al., 2019; Zhang, M. et al., 2022; Zhang, X. et al., 2022; Schmidt et al., 2024), unique post-antennular appendage organization (Zhai et al., 2019a, b, c, 2021; Liu et al., 2021; Schmidt et al., 2021a; O‘Flynn et al., 2024), mouth part characteristics (Liu et al., 2020b), and even brood care features (Ou et al., 2020). More recently, Amira has also contributed to a comprehensive understanding of their morphology (Schmidt et al., 2020, 2021a). Among the species of interest, Leanchoilia illecebrosa stands out as one of the best-studied arthropods of the Chengjiang biota (Hou and Bergström, 1997; Hou et al., 2017). The entire family Leanchoiliidae has a long history of paleontological research (Briggs and Collins 1999; Chen et al., 2004; Liu et al., 2007; Haug et al., 2012). Its remarkable abundance in the Chinese fossil record and exceptional state of preservation makes it an exemplary case for the application of virtual paleontology techniques. These techniques are instrumental in exploring ontogenetic patterns (Liu et al., 2016) and ancestral euarthropod appendage organization (Liu et al., 2021). While specimen YKLP 11439-arthro has yet to be published, specimen YKLP 11422-arthro was previously examined in Liu et al. (2020b), with a particular focus on the organization of its labrum.
We have presented different visualization modes for both specimens using two distinct software programs and have discussed the potential for further processing μ CT-based volumetric data. Both Amira and Drishti offer powerful tools for exploring μ CT-generated volume models. When a quick overview of the scanned specimen’s volume or a general surface representation is sufficient, using volume or isosurface renderings in Amira is more convenient. This is because finding the optimal transfer function in Drishti can often be a time-consuming process. However, Drishti’s unique features include producing organic-looking and highly malleable 3D volumes through painting and carving. Cleaned-up volume models in Drishti, achieved via the morphological operation MOP carve within the Drishti renderer or segmentation in Drishti Paint, can provide clearer perspectives on the fossil, particularly in cases where background scattering was previously problematic. The use of clipping planes in Drishti can unveil hidden internal structures, while Amira’s orthoslices aligned with data slices can highlight different aspects of interest.
Considering the advantages of both programs, we find that general volume models of entire species are best rendered and created with Drishti due to its remarkable flexibility and the realistic and organic appearance of its volumes. For detailed information on specific structures, Amira's volume models may be worth considering, depending on the fossil's quality of preservation. Drishti offers an array of volume exploration modes and represents the superior choice for volumetric model visualizations. Amira may be preferred if quick (iso-) surface models for further mesh processing (Blender, Maya, etc.) are needed. Ultimately, both programs offer high-quality visualization capabilities, and the choice between them may be influenced by personal preferences, experience with the software, and budget considerations, as Amira is a commercial product, while Drishti is open source.
ACKNOWLEDGEMENTS
This study was supported by grants from the Natural Science Foundation of Yunnan Province (202301AS070049, 202401BC070012) to Y.L., who is further supported by the Yunnan Revitalization Talent Support Program, as well as by a Yunnan University PostDoctoral Research Fund (W8223004) to M.S. M.S.´s research is also supported by the Colorful Cloud Postdoctoral Programme of Yunnan Province.
REFERENCES
Briggs, D.E.G. and Collins, D. 1999. The arthropod Alalcomenaeus cambricus Simonetta, from the Middle Cambrian Burgess Shale of British Columbia. Palaeontology, 42:953-977.
https://doi.org/10.1111/1475-4983.00104
Buser, T.J., Boyd, O.F., Cortés, Á., Donatelli, C.M., Kolmann, M.A., Luparell, J.L., Pfeiffenberger, J.A., Sidlauskas, B.L., and Summers, A.P. 2020. The Natural Historian’s Guide to the CT Galaxy: Step-by-Step Instructions for Preparing and Analyzing Computed Tomographic (CT) Data Using Cross-Platform, Open Access Software. Integrative Organismal Biology, 1-25.
https://doi.org/10.1093/iob/obaa009
Cheng, G., Peng, F., Dean, B., and Dong, X. 2011. Internal structure of Cambrian rossil embryo Markuelia revealed in the light of synchrotron radiation x-ray tomographic microscopy. Acta Geologica Sinica, 85(1):81-90.
https://doi.org/10.1111/j.1755-6724.2011.00381.x
Chen, J.‐Y., Waloszek, D., and Maas, A. 2004. A new ‘great appendage’ arthropod from the Lower Cambrian of China and homology of chelicerate chelicerae and raptorial antroventral appendages. Lethaia, 37:3-20.
https://doi.org/10.1080/00241160410004764
Chen, X., Ortega-Hernández, J., Wolfe, J.M., Zhai, D., Hou, X.-G., Chen, A., Mai, H., and Liu, Y. 2019. The appendicular morphology of Sinoburius lunaris and the evolution of the artiopodan clade Xandarellida (Euarthropoda, early Cambrian) from South China. BMC Evolutionary Biology, 19:165.
https://doi.org/10.1186/
Cunningham, J.A., Rahman, I.A., Lautenschlager, S., Rayfield, E.J., and Donoghue, P.C.J. 2014. A virtual world of paleontology. Trends in Ecology and Evolution, 29:347-357.
Frase, T. and Richter, S. 2013. The fate of the onychophoran antenna. Development Genes and Evolution 223:247-251.
https://doi.org/10.1007/s00427-013-0436-x
Gaines, R.R., Hammarlund, E.U., Hou, X.-G., Qi, C., Gabbott, S.E., Zhao, Y., Peng, J., and Canfield, D.E. 2012. Mechanism for Burgess Shale-type preservation. Proceedings of the National Academy of Sciences 109(14):5180-5184.
https://doi.org/10.1063/pt.4.0372
Garwood, R. and Dunlop, J. 2014. The walking dead: Blender as a tool for paleontologists with a case study on extinct arachnids. Journal of Paleontology 88(4):735-746.
https://doi.org/10.1666/13-088
Garwood, R.J., Rahman, I.A., and Sutton, M.D. 2010. From clergymen to computers-the advent of virtual palaeontology. Geology Today, 26:96-100.
Gunz, P., Mitteroecker, P., Neubauer, S., Weber, G.W., and Bookstein, F.L. 2009. Principles for the virtual reconstruction of hominin crania. Journal of Human Evolution 57(1):48-62.
https://doi.org/10.1016/j.jhevol.2009.04.004
Haug, J.T., Briggs, D.E.G., and Haug, C. 2012. Morphology and function in the Cambrian Burgess Shale megacheiran arthropod Leanchoilia superlata and the application of a descriptive matrix. BMC Evolutionary Biology 12:162.
https://doi.org/10.1186/1471-2148-12-162
Hou, X.-G. and Bergström, J. 1997. Arthropods of the Lower Cambrian Chengjiang fauna, southwest China. Fossils and Strata 45:1-116.
https://doi.org/10.18261/8200376931-1997-01
Hou, X.-G., Siveter, D.J., Siveter, D.J., Aldridge, R.J., Cong, P.Y., Gabbott, S.E., Ma, X.-Y., Purnell, M., and Williams, M. 2017. The Cambrian fossils of Chengjiang, China: the flowering of early animal life, 3rd edn. Oxford, UK: Wiley Blackwell.
https://doi.org/10.1002/
Hu, Y., Limaye, A., and Lu, J. 2020. Three-dimensional segmentation of computed tomography data using Drishti Paint: new tools and developments. Royal Society Open Science 7:201033.
https://doi.org/10.1098/rsos.201033
Hu, Y., Limaye, A., and Lu, J. 2024. 3D revisualization: a new method to revisit segmented data. Royal Society Open Science, 11(6):240375.
https://doi.org/10.1098/rsos.240375
Jaszkowiak, K., Keiler, J., Wirkner, C.S., and Richter, S. 2015. The mouth apparatus of Lithodes maja (Crustacea: Decapoda) - form, function and biological role. Acta Zoologica 96:401-417.
https://doi.org/10.1111/azo.12135
Jin, C., Mai, H., Chen, H., Liu, Y., Hou, X.-G., Wen, R., and Zhai, D. 2021. A new species of the Cambrian bivalved euarthropod Pectocaris with axially differentiated enditic armatures. Papers in Palaeontology, 7:1781-1792.
Keiler, J., Richter, S., and Wirkner, C.S. 2015. Evolutionary morphology of the organ systems in squat lobsters and porcelain crabs (Crustacea: Decapoda: Anomala) - an insight into carcinization. Journal of Morphology 276:1-21.
https://doi.org/10.1002/jmor.20311
Keklikoglou, K., Faulwetter, S., Chatzinikolaou, E., Wils, P., Brecko, J., Kvacek, J., Metscher, B., and Arvanitidis, C. 2019. Micro-computed tomography for natural history specimens: a handbook of best practice protocols. European Journal of Taxonomy 522:1-55.
https://doi.org/10.5852/ejt.2019.522
Kudryashev, M., Cyrklaff, M., Alex, B., Lemgruber, L., Baumeister, W., Wallich, R., and Frischknecht, F. 2011. Evidence of direct cell-cell fusion in Borrelia by cryogenic electron tomography. Cellular Microbiology 13(5):731-741.
https://doi.org/10.1111/j.1462-5822.2011.01571.x
Kudryashev, M., Münter, S., Lemgruber, L., Montagne, G., Stahlberg, H., Matuschewski, K., Meissner, M., Cyrklaff, M., and Frischknecht, F. 2012. Structural basis for chirality and directional motility of Plasmodium sporozoites. Cellular Microbiology, 14(11):1757-1768.
https://doi.org/10.1111/j.1462-5822.2012.01836.x
Limaye, A. 2012: Drishti: a volume exploration and presentation tool. Proc. SPIE 8506. Developments in X-Ray Tomography VIII, 85060X.
https://doi.org/10.1117/12.935640
Liu, Y., Hou X.-G., and Bergström, J. 2007. Chengjiang arthropod Leanchoilia illecebrosa (Hou, 1987) reconsidered. Geologiska Föreningens Stockholm Förhandlingar, 129:263-272.
https://doi.org/10.1080/11035890701293263
Liu, Y., Haug, J.T., Haug, C., Briggs, D.E.G., Hou, X.-G. 2014. A 520 million‐year‐old chelicerate larva. Nature Communications, 5:4440.
https://doi.org/10.1038/ncomms5440
Liu, Y., Scholtz, G., and Hou, X.-G. 2015. When a 520-million‐year old Chengjiang fossil meets a modern micro‐CT-a case study. Scientific Reports, 5, 12802.
https://doi.org/10.1038/srep12802
Liu, Y., Melzer, R.R., Haug, J.T., Haug, C., Briggs, D.E.G., Hörnig, M.K., He, Y., and Hou, X.-G. 2016. Three-dimensionally preserved minute larva of a great appendage arthropod from the early Cambrian Chengjiang biota. Proceedings of the National Academy of Sciences, 113:5542-5546.
https://doi.org/10.1073/pnas.1522899113
Liu, Y., Ortega‐Hernández, J., Chen, H., Mai, H., Zhai, D., and Hou, X.‐G. 2020a. Computed tomography sheds new light on the affinities of the enigmatic euarthropod Jianshania furcatus from the early Cambrian Chengjiang biota. BMC Evolutionary Biology, 20:2.
https://doi.org/10.1186/s12862-020-01625-4
Liu, Y., Ortega-Hernández, J., Zhai, D., and Hou, X.-G. 2020b. A reduced labrum in a Cambrian great-appendage euarthropod. Current Biology, 30(15):3057-3061.
https://doi.org/10.1016/j.cub.2020.05.085
Liu, Y., Edgecombe, G.D., Schmidt, M., Bond, A.D., Melzer, R.R., Zhai, D., Mai, H., Zhang, M., and Hou, X.-G. 2021. Exites in Cambrian arthropods and homology of arthropod limb branches. Nature Communications, 12:4619.
https://doi.org/10.1038/s41467-021-24918-8
Lukeneder, S., Lukeneder, A., and Weber, G.W. 2014. Computed reconstruction of spatial ammonoid-shell orientation captured from digitized grinding and landmark data. Computers & Geosciences, 64:104-114.
https://doi.org/10.1016/j.cageo.2013.11.008
Meisslitzer-Ruppitsch, C., Röhrl, C., Ranftler, C., Neumüller, J., Vetterlein, M., Ellinger, A., and Pavelka, M. 2011. The ceramide-enriched trans-Golgi compartments reorganize together with other parts of the Golgi apparatus in response to ATP-depletion. Histochemistry and Cell Biology, 135(2):159-171.
https://doi.org/10.1007/s00418-010-0773-z
O’Flynn, R.J., Williams, M., Yu, M., Gun, J., Auto, D., Schmidt, M., Mai, H., Liu, Y., Edgecombe, G.D. 2024. The early Cambrian Bushizheia yangi and head segmentation in upper stem-group euarthropods. Papers in Palaeontology, 10(3):e1556.
Ou, Q., Vannier, J., Yang, X., Chen, A., Mai, H., Shu, D., Han, J., Fu, D., Wang, R., and Mayer, G.
2020. Evolutionary trade-off in reproduction of Cambrian arthropods. Science Advances, 6:eaaz3376.
https://doi.org/10.1126/sciadv.aaz3376
Ruthensteiner, B. 2008. Soft Part 3D visualization by serial sectioning and computer reconstruction. Zoosymposia, 1:63-100.
https://doi.org/10.11646/zoosymposia.1.1.8
Schmidt, M., Liu, Y., Zhai, D., Hou, X.-G., and Melzer, R.R. 2020. Moving legs: A workflow on how to generate a flexible endopod of the 518-million-year-old Chengjiang arthropod Ercaicunia multinodosa using 3D-kinematics (Cambrian, China). Microscopy Research and Technique, 84(44):695-704.
https://doi.org/10.1002/jemt.23628
Schmidt, M., Liu, Y., Hou, X.-G., Haug, J.T., Haug, C., Mai, H., and Melzer, R.R. 2021a. Intraspecific variation in the Cambrian: new observations on the morphology of the Chengjiang euarthropod Sinoburius lunaris. BMC Ecology & Evolution, 21:127.
https://doi.org/10.1186/s12862-021-01854-1
Schmidt, M., Melzer, R.R., and Bicknell, R.D.C. 2021b. Kinematics of whip spider pedipalps: a 3D comparative morpho-functional approach. Integrative Zoology, 17(1):156-167.
https://doi.org/10.1111/1749-4877.12591
Schmidt, M., Hou, X.-G., Zhai, D., Mai, H., Belojević, J., Chen, X., Melzer, R.R., Ortega-Hernández, J., and Liu,Y. 2022a. Before trilobite legs: Pygmaclypeatus daziensis reconsidered and the ancestral appendicular organization of Cambrian artiopods. Philosophical Transactions of the Royal Society: B-Biological Sciences, 377:20210030.
https://doi.org/10.1098/rstb.2021.0030
Schmidt, M., Melzer, R.R., Plotnick, R.E., and Bicknell, R.D.C. 2022b. Spines and baskets in apex predatory sea scorpions uncover unique feeding strategies using 3D-kinematics. iScience, 25(1):103662.
https://doi.org/10.1016/j.isci.2021.103662
Schmidt, M., Hou, X.-G., Mai, H., Zhou, G., Melzer, R.R., Zhang, X., and Liu, Y. 2024. Unveiling the ventral morphology of a rare early Cambrian great appendage arthropod from the Chengjiang biota of China. BMC Biology, 22:96.
https://doi.org/10.1186/s12915-024-01889-y
Semple, T.L., Peakall, R., and Tatarnic, N.J. 2019. A comprehensive and user-friendly framework for 3D-data visualisation in invertebrates and other organisms. Journal of Morphology, 280(2):223-231.
https://doi.org/10.1002/jmor.20938
Stalling, D., Westerhoff, M., and Hege, H.-C. 2005. Chapter 38. Amira - a highly interactive system for visual data analysis, p. 749-767. In Hansen, C.D. and Johnson, C.R. (eds.), The Visualization Handbook. Elsevier, Amsterdam.
Sutton, M., Rahman, I., and Garwood, R. 2014. Techniques for virtual palaeontology. London: John Wiley & Sons.
https://doi.org/10.1002/9781118591192
Thermo Fisher Scientific 2018. Thermo Scientific Amira Software 6. User´s Guide. 1995-2018 Konrad-Zuse-Zentrum für Informationstechnik Berlin (ZIB), Germany.
https://assets.thermofisher.com/TFS-Assets/MSD/Product-Guides/user-guide-amira-software.pdf
Yin, Z. and Lu, J. 2019. Virtual Palaeontology: when fossils illuminated by X-ray. Palaeoworld, 28:425428.
Zhai, D., Edgecombe, G.D., Bond, A.D., Mai, H., Hou, X.-G., and Liu, Y. 2019a. Fine-scale appendage structure of the Cambrian trilobitomorph Naraoia spinosa and its ontogenetic and ecological implications. Proceedings of the Royal Society-B: Biological Sciences, 286(1916).
https://doi.org/10.1098/rspb.2019.2371
Zhai, D., Ortega-Hernández, J., Wolfe, J.M., Hou, X.-G., Cao, C., and Liu, Y. 2019b. Three-dimensionally preserved appendages in an early Cambrian stem-group pancrustacean. Current Biology, 29(1):171-177.
https://doi.org/10.1016/j.cub.2018.11.060
Zhai, D., Williams, M., Siveter, D.J., Harvey, T.H.P., Sansom, R.S., Gabbott, S.E., Siveter, D.J., Ma, X., Zhou, R., Liu, Y., and Hou, X.-G. 2019c. Variation in appendages in early Cambrian bradoriids reveals a wide range of body plans in stem-euarthropods. Communications Biology 2:329.
https://doi.org/10.1038/s42003-019-0573-5
Zhai, D., Williams, M., Siveter, D. J., Siveter, D. J., Harvey, T. H. P., Sansom, R. S., Mai, H., Zhou, R., and Hou, X.-G. 2022. Chuandianella ovata: An early Cambrian stem euarthropod with feather-like appendages. Palaeontologia Electronica, 25(1):a7.
https://doi.org/10.26879/1172
Zhang, M., Liu, Y., Hou, X.-G., Ortega-Hernández, J., Mai, H., Schmidt, M., Melzer, R.R., and Guo, J. 2022. Ventral morphology of the non-trilobite artiopod Retifacies abnormalis Hou, Chen and Lu, 1989, from the early Cambrian Chengjiang Biota, China. Biology, 11(8):1235.
https://doi.org/10.3390/biology11081235
Zhang, X., Liu, Y., O’Flynn, R., Schmidt, M., Melzer, R.R., Hou, X.-G., Mai, H., Guo, J., Yu, M., and Ortega-Hernández, J. 2022. Ventral organization of Jianfengia multisegmentalis Hou, and its implications for the head segmentation of megacheirans. Palaeontology, 65(5):e12624.
https://doi.org/10.1111/pala.12624
Zhao, T., Hou, X.-G., Zhai, D., Wu, D., Chen, H., Zhang, S.-G., and Liu, Y. 2017. Application of the micro-CT. technique in the studies of arthropods from the Chengjiang biota: a case of Misszhouia longicaudata. Acta Palaeontologica Sinica, 56(4):476-482.

